Synthesis, Characterizations, Biological, and Molecular Docking Studies of Some Amino Acid Schiff Bases with Their Cobalt(II) Complexes ()
1. Introduction
The field of Schiff base complexes has been fast developed because of the wide possible structures for the ligands depending upon the aldehydes and amines considered [1] [2] .
Various Schiff bases were reported to possess genotoxicity [3] , antibacterial [4] , and antifungal activities [5] . The increasing interest in transition metal complexes containing a Schiff base ligand is derived from their well-established role in biological systems as well as their catalytic and pharmaceutical applications [6] . In other hand, Schiff base and their metal complexes have been characterized because of their interesting and important properties as the ability of bind toxic and heavy metal atoms [7] , undergoes tautomerism [8] , and exhibits catalytic reduction [9] , and photochromism [10] . The role of the metal Schiff bases complexes in such applications, is related to molecular structure, thus, it is quite important to have a good understanding of the structure of such metal complexes.
Today, Schiff base have been used extensively as ligands in the field of coordination chemistry, because of intermolecular hydrogen bonds between the “O” and the “N” atoms, which play an important role in the formation of metal complexes and the proton transfer from the hydroxyl “O” to the imine “N” atoms [2] . In this respect, complexes of metal ions with Schiff base derived from salicylaldehyde and amino acids have been synthesized and characterized by using different physical techniques and coordination geometry structures were proposed based on their results.
The aim of the present paper is to synthesize Schiff bases which expected to have biological effect which derived from the reaction of salicylaldehyde with three amino acids (valine, leucine, and isoleucine). In many cases the bioactivity of Schiff base increases upon coordination to metal ions. Therefore, it would be of interest to describe the divalent cobalt(II) complexes of the synthesized Schiff bases.
2. Experimental
2.1. Materials and Reagents
All material used in the investigation were of reagent grade and were purchased commercially from British Drug House (BDH) and used as received. They include amino acids (valine, leucine, and isoleucine), salicylaldehyde, Cobalt (II) chloride, absolute methanol and ethanol. Double distilled water was used during the experimental process.
2.2. Preparation of Schiff Base
Schiff base (1-valine, 2-leucine, and 3-isoleucine) under investigation were prepared according to the procedure previously described in the literature [11] , Thus, in a 50 ml conical flask, prepare a separate solution by dissolving (1.2 g) of salicylaldehyde (0.75 g) of amino acids (1 - 3) and (0.4 g) of sodium hydroxide, in 15 ml of ethanol. Mix the salicylaldehyde solution within the proper amino acid solution. The mixture was stirred for 8 hours on a water bath at 70˚C. The obtained precipitate was filtered off and washed with cold ethanol. The solid products were recrystallized from hot ethanol to give yellow crystals, dried in vacuo over silica gel. The purity of the prepared compounds was checked by elemental analysis, IR, 1H NMR spectra.
2.3. Preparation of Metal Complexes
The preparation of the metal complexes under study were prepared according to procedure previously described in the literature [12] , thus, the complex [CoL2]nH2O have been prepared by the reaction of (0.590 gm) (2 mmole) of ligands (1 - 3) with (1 mmole) of Cobalt(II) chloride CoCl2∙6H2O (0.238 gm) dissolved in (20 mls) absolute ethanol and stirring for 24 hours. The obtained complexes were collected after evaporation of ethanol and was left in the desiccator to be dried under P2O5.
2.4. Measurements
The prepared Schiff base and their metal complexes were provide by elemental analysis (C%, H%, N%) which were performed at the microanalytical center, Cairo University, the analysis were performed twice to check the accuracy of the analysis data. The IR spectra were recorded on as 8001-PC FTIR Shimadzu spectrophotometer using KBr pellets. Mass spectra of the complexes were performed using GC-2010 Shimadzu at Microanalytical center, Cairo University, Giza, Egypt. The magnetic susceptibility measurements for the complexes were determined by the Gouy balance using Hg[Co(SCN)4] as calibration. 1H NMR and 13C NMR were recorded on a Bruker DPX-400 spectrometer using DMSO d6, as the solvent and TMS as internal standard. Molar conductivity of 10−3 M solutions of the complexes in DMF was measured on the conductivity meter ORION model 150 of 0.6 cell constant.
3. Results and Discussions
The analytical data for the isolated complexes indicates that the Cobalt(II) complexes ions under investigation are of 1:2 ratio (M2+: ligand). The molar conductance of the free synthesized ligands reading values indicating that the free ligands are electrolytic nature by the formation of the monosodium salts.
The reaction of Schiff base ligands (1-valine, 2-Leucine, and 3-Isoleucine) with the Cobalt(II) ions under investigation give a type of reactions which can be represented by the following equations:
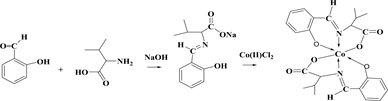 (1)
(1)
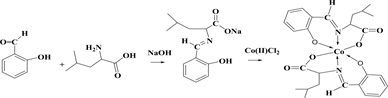 (2)
(2)
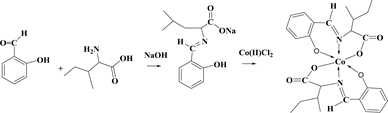 (3)
(3)
Chemical equations of synthesis, where 1: valine amino acid, 2: leucine amino acid 3: isoleucine amino acid.
3.1. Microanalysis Data
The CHN elemental analysis data of the investigation Schiff base ligands (1 - 3) and its Cobalt(II) metal complexes as shown in Table 1, exhibit the metal complexes formation of [ML2] ratio. It was found that the theoretical values are in a good agreement with the found data.
3.2. Infrared Spectral Studies
The characteristic IR absorption bands of the studied amino acid Schiff bases and their Cobalt(II) complexes are presented in Table 2. The IR spectra of Schiff bases under study illustrate the following stretching frequencies:
1) A broad stretching frequency range between (2959 - 2922 cm−1) in case of the free ligands which confirm OH group with shifting effect in case of complexes indicating the coordination of “O” group in complexation. The spectra of the Co(II) complexes exhibit broad bands in the range of (3415 - 3354 cm−1) which analogous to the presence of hydrated water molecules [13] .
2) A strong to medium intensities bands are assigned to carbon nitrogen double bond (H-C=N) with stretching frequencies range between (1633 - 1625 cm−1). The shifting of these bands to low frequencies comparing to the Schiff base ligands confirmed the participation of this group in complexation through Nitrogen atom [14] .
3) A strong to medium intensity bands are assigned to carbonyl group in Schiff base γ (C=O). They have stretching frequencies range between (1610 - 1593 cm−1)
4) Schiff base show a linkage for sodium of carboxyl acid, (O-Na) at stretching range between γ (755 - 753 cm−1). These values confirm the existence of these compounds as a sodium salts for carboxylic acids. The bands appeared at stretching frequencies range between γ (1522 - 1462 cm−1) are assigned for (C=C).
The appearance of new bands in the range γ (563 - 461 cm−1) [15] , and (421 - 277 cm−1) [16] can be assigned to γ (M-O) and v (M-N) stretching vibrations observed in the metal complexes and absent in the free Schiff base ligands provide evidence that (C=O) and (C=N) moiety could be bonded to the metal ion through the Oxygen and the Nitrogen atoms.
![]()
Table 1. Physical properties and analytical data of amino acid Schiff base with its metal complexes.
![]()
Table 2. FTIR spectra bands and electronic spectra bands of Amino Acid Schiff Base and Cobalt(II) Complexes using KBr pellets (cm−1), and nm (cm−1) for uv bands.
3.3. Electronic Spectra Studies
The electronic spectral results of the studied Schiff bases (1 - 3) with Cobalt(II) complexes are presented in Table 2. The electronic spectra studies were carried out in DMF solvent. The electronic spectra of the Schiff bases (1 - 3) show a low energy bands at (413 - 410 nm) which assigned to NH form, while (314 - 312 nm) which assigned to the OH form are in agreement with the NMR results Table 3. For electronic spectra of the Schiff bases with Cobalt(II) complexes, both NH and OH peaks are not assigned with agreement to the occurrence of Schiff bases (1 - 3) metal complexes, as it is illustrated in Table 2. The electronic spectra of the Schiff base exihibit Π to Π* (phenyl ring) and n to Π* (H-C=N) transition [17] . Whereas, for Co(II) metal complexes, the electronic spectral results display two bands for Val-Co at 360 nm (27777 cm−1), and 727 nm
![]()
Table 3. 1H NMR chemical shift (ppm) and 13C NMR chemical shift (ppm) in DMSO Deuterium solvent at 25˚.
(13755 cm−1), assigned to 4T1g(F) to 4T2g(F), and 4T1g(F) to 4T1g(p), transition in octahedral field [13] , indicating an octahedral geometry around Co(II) ion. Their magnetic moments at room temperature (μeff = ~4.4 μB), corroborate with high-spin octahedral coordination of the Co(II)ion [15] [18] .
3.4. 1H NMR Spectra
The 1H NMR spectra of the investigated Schiff base ligands (1 - 3) presented in Table 3, and the assignment clearly presented as it numbered in Figure 1, which showed the following frequencies.
・ A singlet at (14.50 - 14.30 ppm) corresponds to phenolic -OH proton [19]
・ A sharp singlet frequency at (8.43 - 8.35 ppm) was observed and attributed to the α-H, for CH=N group Table 3.
・ The multiplets observed in the region (7.35 - 6.70 ppm) are ascribed to the aromatic hydrogen.
・ The triplet observed in the region (3.65 - 3.52 ppm) are assigned for (HC-R) group as illustrated in Table 3.
3.5. 13C NMR Spectra
The 13C NMR spectra of the investigated Schiff base ligands (1 - 3) presented in Table 3, and the assignment clearly presented as it numbered in Figure 1, which showed the following frequencies:
・ The chemical shift values for C1, C3, C5 of the aromatic carbons observed symmetrically in the range (118.05 - 116.15 ppm), where the chemical shift values for the other carbon atoms of the aromatic ring C4, C6 are observed at the range of (131.93 - 131.24 ppm) and the C2 have the chemical shift values in the range (164.76 - 163.26 ppm) influenced by the phenolic group.
・ The chemical shift value for α-C (H-C=N) is observed at (163.45 ppm) influenced by imine group.
・ The chemical shift value for δC1− (R-CH) for valine, leucine, and Isoleucine are observed at (78.05, 70.45, and 75.43 ppm) respectively as in Table 3.
・ The carboxylic carbon δC2− have the chemical shift value in the range (173.72 - 173.00 ppm) influenced by the oxygen atoms of the carboxylic acid group.
<
![]()
Figure 1. Amino acid Schiff bases (1 - 3) (1) R = CH(CH3)2, (2) R = CH2CH(CH3)2, (3) R = CH(CH3)CH2CH3.
13C NMR spectrum of the Schiff base ligands were recorded in DMSO d6, all assignments of the carbon atoms were founds on the expected regions [19] .
3.6. Mass Spectra of the Schiff Base Ligands (1-3) and Their Co(II) Metal Complexes
The mass spectral fragmentation of the Schiff base ligands (1 - 3) and Co(II) metal complexes are shown in Table 4, as follows:
1) Valine Schiff base ligand is represented by the peak at m/e+ = 243 analogous to [C12H14NO3Na] structure. Another peak at m/e+ = 221 is equal to the formula [C12H14NO3]. The structure [C12H14NO] is analogous to peak at m/e+ = 176. Meanwhile, the peak at m/e+ = 132 is equal to the formula [C9H9NO], where the last peak at m/e+ = 107 is attributing to [C7H6O].
2) Leucine Schiff base ligand is represented by peak at m/e+ = 257 analogous to [C13H16NO3Na]. The peak at m/e+ = 242 is analogous to the loss of methyl molecule from the ligand, where the loss of another methyl molecule give a peak at m/e+ = 227. The peak at m/e+ = 202 is analogous to the loss of sodium atom. Meanwhile, the last peak at m/e+ = 132 is attributing to formula [C8H7NO].
3) Isoleucine Schiff base ligand of the formula [C13H16NO3Na] is represented by a base peak at m/e+ = 257, due to original molecular weight. The peak at m/e+ = 242 is analogous to the loss of the methyl molecule (
) from the original ligand. The loss of another (
) gives a peak at m/e+ = 227. Where the loss of sodium atom gives a peak at m/e+ = 202. The same spectrum shows a peak at m/e+ = 132 corresponding to the loss of (C3H3-O2), from the free ligand.
4) Valine Schiff base Co(II) metal complex:
The spectrum of Schiff base Co metal complex shows a molecular ion peak at m/e+ = 497, corresponding to [(C12H14NO3)2Co]. Whereas, the peak at m/e+ = 414 analogous to [(C9H7NO2)2Co] as a result of a loss of (C3H7)2. The peak at m/e+ = 384 due to [(C9H6NO2)2Co]. Whereas, the peak at m/e+ = 369 analogous to [(C8H6NO2)2Co] as result of loss of (
). Another loss of (C2H4) gives a peak at m/e+ = 339. The peak appeared at m/e+ = 313 attributed to [(C7H5NO2)2Co].
5) Leucine Schiff base Co(II) metal complex:
The spectrum shows a molecular ion peak at m/e+ = 525 corresponding to [(C13H16NO3)2Co]. The peak at m/e+ = 468 is analogous to [(C9H8NO3)Co] as a loss of (C4H9). Whereas, the peak at m/e+ = 412 is a result of a loss of another
![]()
Table 4. Mass spectral fragmentation of Schiff base (1 - 3) and their Co(II) metal complexes.
(C4H9). The peak at m/e+ = 380 is due to loss of two oxygen atoms. Whereas, the peak at m/e+ = 355 is a result of loss of (C2H4) from the original complex.
6) Isoleucine Schiff base Co(II) metal complex:
The spectrum shows a molecular ion peak at m/e+ = 597 corresponding to [(C13H16NO3)2Co∙4H2O]. The loss of 4 water molecules resulting a peak at m/e+ = 525, due to the original complex [(C13H16NO3)2Co]. The peak at m/e+ = 468 is a result of a loss of (C4H9) from the original peak. Whereas, a loss of another (C4H9) resulting a peak m/e+ = 412, due to the molecular formula [(C9H8NO3)2Co]. The peak at m/e+ = 380 is analogous to the loss of two oxygen atoms which is attributed to formula [(C9H8NO2)2Co]. the last peak at m/e+ = 355 corresponding to a loss of (C2H4) from the complex.
3.7. Biological Activities
Antibacterial and antifungal activity of the synthesized ligands and their cobalt complexes are listed in Table 5. The results, displayed in Figure 2, are summarized in the following:
1) The all free ligands are biologically active against gram positive bacteria (Staphylococcus aureus and Bacillus subtilis) and gram negative bacteria (Escherichia coli and Pseudomonas aeruginosa), their Co(II) complexes show more bacterial activity against most bacterial species.
2) The three ligands are inactive against fungi (Candida albicans and Aspergillus flavus) but their complexes show activity against Candida albicans.
These results may be explained onthe bases that the metal complexes penetrate more easily through the bacterial cell wall, due to the proteic denaturation of the sulphydrile group [20] , destroying the bacterial cell wall.
3.8. Molecular Docking Studies
The molecular docking approach can be used to model the interaction between a small molecule and a protein at the atomic level, which allow us to characterize the behavior of small molecules in the binding site of target proteins as well as to elucidate fundamental biochemical processes [18] . The docking process involves two basic steps: prediction of the ligand conformation as well as its position and
![]()
Table 5. Antibacterial and antifungal activities of the free ligands and their dimeric Co (II) complexes.
![]()
Figure 2. Biological activities of the free ligands and their cobalt(II) complexes towards different types of bacteria and fungi.
orientation within these sites (usually referred to as pose) and assessment of the binding affinity. These two steps are related to sampling methods and scoring schemes, respectively, which will be discussed in the theory section [21] Mol format file was used for docking process. Docking of these compounds into ATP binding sites of 3GEY transferase enzyme was performed using Moe 2009 program to stimulate the binding model and the protein crystal structure was obtained from the Protein Data Bank (PDB).
Molecular docking investigation proved that, ligands and their Co2+ complexes have shown interesting interaction with active site amino acids of ribosyltransferase (code: 3GEY). All ligands have many interactions as pointed in (Figure 3). For L1, the presence of side chain acceptor interaction from the Tyr 582 to the carbonyl carboxylic group of the igand. For L2 and L3 the presence of metal contact receptor interaction between the nitrogen of His A537 and the sodium carboxylate. For Co (II) complexes the presence of polar and arene-arene type of bonding with Tyr A582.
![]()
Figure 3. Docking results of ligands and Co(II) complexes.
4. Conclusion
The Schiff bases derived from the reaction of amino acid (1 - 3) with salicylaldehyde have been synthesized. The satisfactory analytical data and the spectroscopic studies suggest the complexes are of the general formula [ML2], where M is Co(II). According to magnetic measurements, electronic spectra, and IR data, the Schiff base ligands coordinated to the metal atom through hydroxyl oxygen atom of, imino nitrogen and phenolic oxygen. The electronic spectra of the metal complexes and their magnetic susceptibility measurements suggest octahedral structure is the probable coordination geometry for the investigated complexes. The geometrical structures concerning the complexes are proposed in the chemical equations cited above. Their Co(II) complexes show more bacterial activity against most bacterial species and Candida albicans fungus.