Molecular Study of a New Disease of Peach in Iran Associated with a Phytoplasma ()
1. Introduction
Peach (Prunus persica) is one of the most important fruit trees around the world including Iran. Kurdistan is a major peach growing province in the country with a total production of 3732 tones per year [1]. Stone fruits such as peach are susceptible to several diseases associated with phytoplasmas. In Europe, European stone fruit yellows (ESFY) phytoplasmas cause apricot (Prunus armeniaca) chlorotic leaf roll, decline and leptonecrosis of Japanese plum (Prunus salicina), yellows and decline of peach, on European plum (Prunus domestica) and almond (Prunus dulcis) [2,3]. Other diseases in stone fruits are peach X-disease phytoplasmas, peach yellow leaf roll phytoplasma, and cherry lethal yellows phytoplasma [4].
Recently, “Candidatus Phytoplasma phoenicium” has been reported in association with a phytoplasma diseases on nectarine and peach trees in south of Lebanon and RFLP analysis of nested PCR products of 16S rDNA demonstrated that this phytoplasma belonged to the pigeon pea witches’ broom group (16SrIX) and blast analysis of over 1000 nucleotides showed the presence of “Candidatus Phytoplasma phoenicium” in infected trees [5]. In 2004, it was reported that an unknown phytoplasma was associated with yellows disease in peach trees in Kurdistan Province of Iran [6]. Results of some preliminary studies in recent years have shown that “Candidatus Phytoplasma aurantifolia”, “Ca. Phytoplasma solani” and “Ca. Phytoplasma trifolii” were prevalent in peach growing areas in the central and north-west regions of Iran [7]. During a survey for detection of stone fruit phytoplasma disease in Kurdistan Province of Iran, symptoms including shoot proliferation, yellowing and leaf growth retardation were observed on Peach (Prunus persica [L.] Batsch) trees in Sanandaj orchards. A phytoplasma like agent from the above mentioned province has been successfully transmitted from peach to peach and almond by grafting.
The objective of this study was to identify and confirm the causal agent of peach yellows disease in Kurdistan province of Iran using molecular techniques.
2. Materials
Collection and Propagation of Phytoplasma
Symptomatic peach samples were obtained from naturally infected orchards in Sanandaj area of Kurdistan province. All samples consisted of twigs with foliage; Phytoplasmas from some source plants were transmitted by grafting to potted peach and almond young trees. All pots were kept in a screened greenhouse with periodic insecticide treatments to prevent introduction or transmission of unwanted agents.
3. Methods
3.1. DNA Extraction and Nested PCR
The nucleic acid extraction procedure described by Kollar et al. (1990) [8] was followed in this study. DNA concentrations were estimated by using a spectrophotometer according to standard techniques and procedures [9].
Phytoplasmal rDNA was amplified in either single or nested PCR reactions. Four pairs of primers were used to obtain products as follows: P1 and P7 [10,11] for 1.8 kb comprising nearly the entire 16S rDNA gene and all of the 16S-23S spacer regions; R16F2 and R16R2 [12] for 1.2 kb of 16S rDNA. PCRs with primer pairs P1/P7 and R16F2/R2 were performed as previously described [4, 12]. In 25 µl of mixtures containing 100 ng nucleic acid and 1.25 U Taq DNA polymerase with the following concentrations of other reagents: each deoxynucleoside triphosphate, 200 µM; each primer, 0.4 µM, 1× Buffer II (Perkin-Elmer); and MgCl2, 3.5 mM. PCR amplifications were performed using an automated thermocycler (Biorad DNA Thermal Cycler). Parameters used for 35-cycle PCR with primer pair P1/P7 were denaturation at 94˚C for 30 s (5 min for the first cycle), annealing for 55 s at 58˚C, and primer extension for 90 s (10 min for the last cycle) at 72˚C. Parameters used with primer pairs R16F2/ R2 and R16mF2/R16mR1 for 35-cycle PCR were denaturation at 94˚C for 1 min (2 min for the first cycle), annealing for 2 min at 56˚C, and primer extension for 3 min (10 min for last cycle) at 72˚C.
PCR amplifications were performed using an automated thermocycler (Biorad DNA Thermal Cycler). PCR products (5 µl) were detected and their sizes were estimated by electrophoresis with size standards in 1% agarose gel in Tris Borate EDTA (TBE) buffer, followed by staining with ethidium bromide and visualization of the DNA bands using an UV transilluminator.
3.2. RFLP Analyses
Variability among phytoplasmas in 16S rDNA and in the 16S-23S spacer were studied by digesting aliquots of the PCR products obtained with primers R16F2 and R16R2 of the strains. Aliquots of the PCR products obtained with primer pair R16F2 and R16R2 from positive samples were digested at 37˚C overnight by using restriction enzyme AluI, EcoRI, HaeIII, HinfI, HpaI, MboI, MseI, RsaI and TaqI (Fermentase) at 65˚C overnight. For each digestion, 10 U enzymes were used. The restriction products were separated by electrophoresis in a 2% Agarose gel prepared in TBE [9] and stained with ethidium bromide. DNA bands were visualized as described above.
3.3. Nucleotide Sequencing and Phylogenetic Analyses
The nested PCR amplimers of peach strains were sequenced. The PCR products DNA concentrations were estimated spectrophotometrically and adjusted for sequencing using sterile distilled water. Sequencing was performed using automatic DNA sequencer (ABI Prism® Big Dye™ Terminator Cycle Sequencing (Applied Biosystems, USA)) in Abosina research Center. Blast analysis was performed using sequences available at National Center for Biotechnology Information (NCBI).
Cladistic analyses of the alignments (approximately 1200 bp) were carried out with Clastal x version 1.81 software and the phylogenetic tree was constructed by parsimony analyses. Acholeplasma lowdwiki was selected as the out-group to root the tree. The stability of relationships was assessed by performing bootstrap analyses in 100 resembling. The nucleotide sequences of peach yellows phytoplasma was deposited in GeneBank as accession numbers GU338000.
4. Results
4.1. Visual Observations in the Field
During the surveys conducted in the late summer of 2010, peach trees showed leaf growth retardation and little leaves (Figure 1). The samples that were isolated from peach trees showing yellows symptoms were successfully transmitted from peach to peach and almond and grafted peach and almonds showed yellowing symptoms 4 - 6 months after grafting (Figure 2).
4.2. Detection of Phytoplasmas in Diseased Plants
A nested PCR with primers R16F2n/R16R2 followed by

Figure 1. Yellowing symptoms observed usually associated with phytoplasma infections.

Figure 2. Grafted peach (left) and almond (right) show proliferation and yellowing symptoms 5 - 6 months after grafting.
primary PCR by P1/P7 detected phytoplasma DNA. An approximately 1250 bp fragment, in 20 out of 35 diseased peach plants were tested. No amplification was obtained from healthy plants or water control. All nested PCR positive samples were showed yellowing symptoms in orchards. Nested PCR on grafted peach and almond samples also were positive.
4.3. RFLP Analyses of Amplified 16S rDNA
The RFLP patterns of all detected phytoplasma isolates were repeatedly shown to be identical (Figure 3). According to RFLP analysis of 16S rRNA genes for identification of phytoplasma groups [13], the restriction site pattern of the peach infecting phytoplasma isolates were similar to “Candidatus Phytoplasma phoenicium”.
4.4. Nucleotide Sequence and Phylogenetic Analyses
The amplified product of 1250 bp of the primer pair R16r2n/F2 from peach infecting phytoplasma were se-

Figure 3. (A) From left to right: phytoplasma 16S rDNA sequences (nested-PCR products amplified using primer pair (R16F2n/R16R2), Gene Ruler 1 Kb DNA Ladder, RFLP analyses of phytoplasma 16S rDNA sequences (nested-PCR products amplified using primer pair (R16F2n/ R16R2) from Kordestane isolate 26, 29, 6 and Fars almond respectively with MspI (wells 3 - 6); (B) From left to right: Gene Ruler 1 Kb DNA Ladder, phytoplasma 16S rDNA sequences (nested-PCR products amplified using primer pair (R16F2n/R16R2), RFLP analyses of phytoplasma 16S rDNA sequences (nested-PCR products amplified using primer pair (R16F2n/R16R2) from Kordestane isolate 26, 29, 6 and respectively with Bsp143I (wells 3 - 6), (C) From left to right: Gene Ruler 1 Kb DNA Ladder, RFLP analyses of phytoplasma 16S rDNA sequences (nested-PCR products amplified using primer pair (R16F2n/R16R2) from Kordestane isolate 26, 29, 6 and Fars almond respectively with RsaI (wells 2 - 5).
quenced and results have been deposited in the GenBank database (Accession No. GU338000). Blast searches in GeneBank were performed in phytoplasmas 16S rRNA gene (about 1240 bp). Results showed that the peach phytoplasma isolate PP6 obtained from Kurdistan was closely related to Almond witches’-broom phytoplasma (GeneBank accession no. AF390136, AF455041, AF455040, AF455038, AF455039, AF515636, AF515637) with 98% identity. The isolate PP6 was closely related to Pigeon pea witches’-broom phytoplasma (GeneBank accession no. EF186825). The phylogenetic tree was constructed by parsimony analyses of nearly identical length of 16S rRNA gene from peach phytoplasma isolate 6, 29 representative phytoplasmas, Acholeplasma laidlawii (Figure 4). The tree showed that the isolate Peach 6 and almond whitches-broom phytoplasma clustered together and they belong to pigeon pea witches-broom group.
5. Discussion
Stone fruits have been reported to be affected by several diseases associated with plant pathogenic phytoplasmas including European stone fruit yellows or apricot chlorotic leaf roll phytoplasma [2,14], peach X-disease phy-
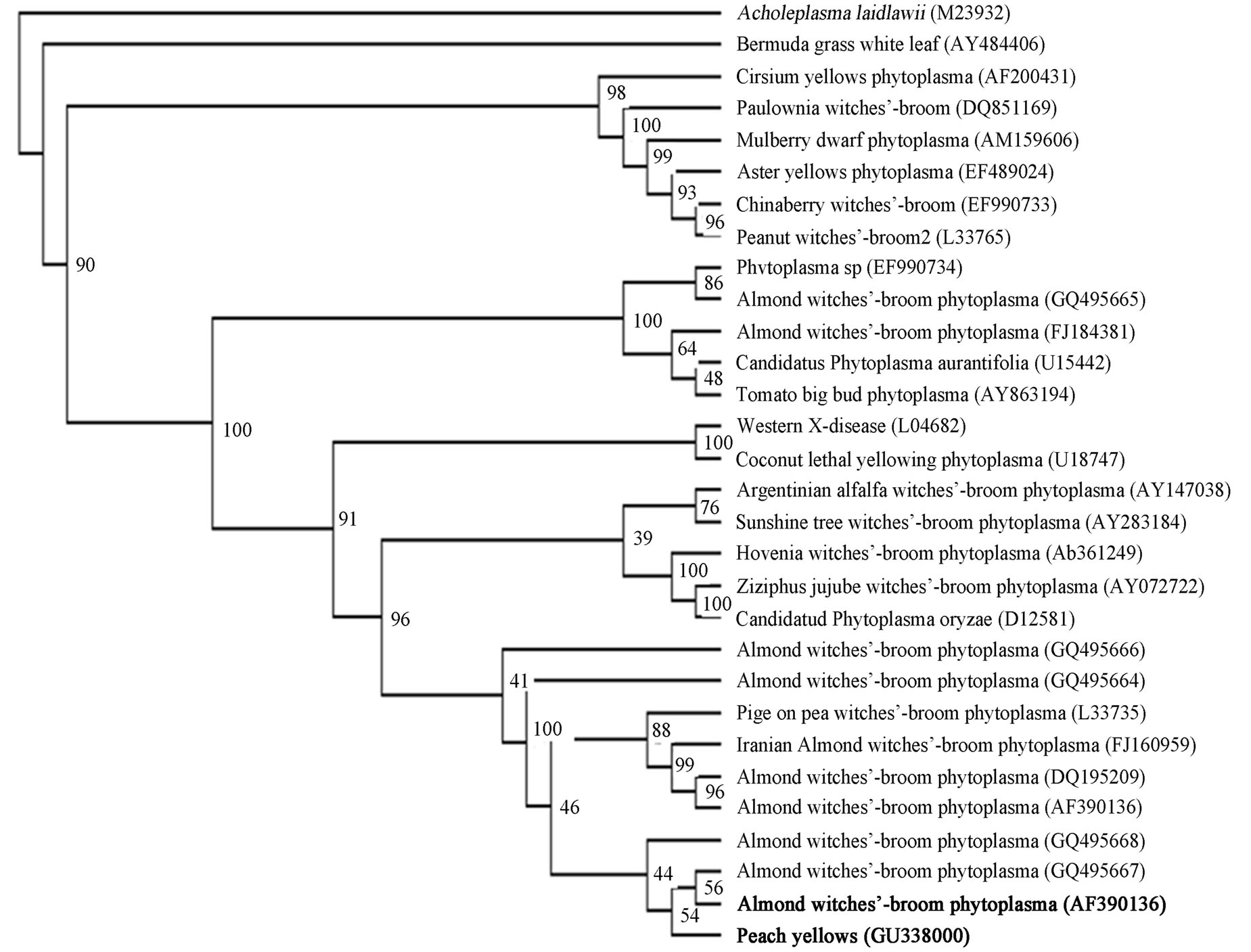
Figure 4. A phylogenetic tree was constructed by parsimony analyses, represents the evolutionary relationships of 29 phytoplasma species based on the 16 S ribosomal protein gene by using the Maximum Parsimony method. Acholeplasma laidlawii (M23932) was used as outgroup. 16 S ribosomal protein gene sequence accession numbers are shown in parentheses. Position of Peach yellows phytoplasma (GU338000) and Almond witches broom phytoplasma (AF390136) respect to other reference phytoplasmas downloaded from GenBank are shown as phytoplasma names that are in bold. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) are shown next to the branches.
toplasmas (eastern-X or western-X diseases), peach yellow leaf roll phytoplasma, cherry lethal yellows phytoplasma [4] and “Ca. Phytoplasma phoenicium” [15,16].
In this study, we showed that a phytoplasma is associated with peach yellows symptoms in Kurdistan province of Iran that showed 98% sequence homology with almond witches’ broom phytoplasma, “Ca. Phytoplasma phoenicium” in blast analysis. Phylogenetic trees constructed from 16S sequences are in total agreement with the major groups previously established by full-length sequences of 16S rRNA gene [4].
Almond witches’ broom has been reported as a widespread and devastating disease in Iran and Lebanon [16, 17]. It can spread rapidly and kill trees at any age within 2 - 3 years [17] and transmitted by dodder to peach and nectarine trees in greenhouse condition [16,17]. We have previously reported that almond witches broom phytoplasma is present in all almond important growing area in Iran [18]. The wide spread of Almond witches’ broom over large geographical areas in Iran also suggest the presence of an efficient insect vector.
The vector of Almond witches’ broom has not been identified yet in Iran, but it is possible that this phytoplasma is transmitted from almond to peach trees by insect vector or grafting. Our results also show that samples with proliferation symptoms have not been able to be transmitted from symptomatic peach to healthy almond and peach by grafting. The nested PCR amplification using primer pair R16F2n/R16R2 for detection of phytoplasma in these samples were negative. These results disagree with those obtained by Abou-Jawdah et al in Lebanon [5].
6. Conclusion
Based on the overall results of our study, it may be concluded that different strain of “Ca. Phytoplasma phoenicium”, could be the cause of disease in different peach cultivars resulting in the appearance of different symptoms when infected by almond witches broom phytoplasma. Further investigation, however, should be conducted on the “Ca. Phytoplasma phoenicium” isolates that cause disease in peach trees.