1. INTRODUCTION
Autoimmune diseases can affect different organs and tissues [1,2]. In these conditions, the breakdown in the balance between autoregulatory immune pathways and pathogenic autoreactivity leads to aggressive antibody and T-cell mediated reactions directed against antigens expressed by the host’s own tissues [2]. It is recognised that a complex interaction of genetic/environmental factors underlies the etiopathogenesis of autoimmune disorders [1,3]. Their incidence is increasing worldwide [2]. This has stimulated investigations on their etiopathogenesis. With the advent of new tools, such as genome wide association studies (GWAS) [4], the evidence was provided for new susceptibility loci and candidate genes in the disease process, involved in the immunological synapse and T cell activation [5]. Among organ-specific autoimmune disorders, insulin-dependent diabetes mellitus [Type 1 diabetes mellitus, (T1D)] is a polygenic autoimmune disease, which occurs in human leukocyte antigens (HLAs) genetically predisposed individuals as a consequence of organ-specific immune destruction of the insulin-producing β cells in the islets of Langherans within the pancreas [6] by autoreactive T cells. Regarding T1D, currently 15 loci are shared with other immune-mediated disorders [7-11]. Among the identified candidates [7], there are: cytotoxic T lymphocyte-associated antigen 4 (CTLA4) which suppresses T cell activaTion; protein tyrosine phosphatase non-receptor type 22 (PTPN22) affecting the T cell receptor signaling pathway [8]; the interleukin-2 receptor alpha (IL-2Rα)/CD25 gene [9] playing a pivotal role in the development and function of regulatory T cells (Tregs), IL-7R [10] and SH2B adaptor protein 3 (SH2B3) [11].
In recent years, a special focus was put on genes controlling the proteolytic pathways involved in antigen presentation [12]. In the autoimmune process, immune responses are generated against self-peptides which are presented in the context of Major Histocompatibility Complex (MHC) class I molecules. Antigenic peptides are produced in the cytoplasm as proteolytic intermediates by degradation of endogenous proteins; this occurs through proteases including the multicatalytic proteasome [13]. The transporters associated with antigen processing (TAP1 and TAP2) transport proteolytic intermediates into the endoplasmic reticulum (ER) for further processing by ER aminopeptidases, ERAP1 and ERAP2 (ERAP1/2), before being loaded onto MHC class I molecules [14,15]. The highly polymorphic molecules MHC class I [16] represent primary components of host immune response, therefore, their genetic linkage with several autoimmune disorders was investigated [15]. GWAS and follow-up case-control studies were conducted leading to ERAP1/2 single nucleotide polymorphisms (SNPs) identification in several autoimmune conditions [15], suggesting a putative role in their pathogenesis with associated alleles through altered MHC class I peptide presentation. Close linkages have been found in a number of diseases, including ankylosing spondylitis (AS) [17-19], multiple sclerosis (MS) [20], Crohn’s disease (CD) [21] and T1D [22].
Recently Fung and colleagues [22] selected SNPs for genotyping that had shown convincingly associated with at least one autoimmune condition within the previous two years. Among the others, the evidence for some associations with T1D was also found in the region containing 5q15/ERAP1 (rs30187, ARTS1). ERAP1 was also tested for interaction with HLA class I, especially with HLA-A and HLA-B, but no evidence for an interaction was observed by the authors [22].
In the light of the foregoing, we conducted a case-control genetic association study to test the association of ERAP1 polymorphism rs30187 (minor A allele) with the development of T1D in an Italian sample of Caucasian ethnicity.
2. MATHERIALS AND METHODS
2.1. Subjects
The patient group consisted of 234 T1D patients who were referred from the Department of Endocrinology at Bambino Gesù Children’s Hospital (OPBG) during the past 10 years. Patient’s sera were tested for autoantibodies (Abs) to glutamic acid decarboxylase (isoform 65) (GADA), protein tyrosine phosphatase IA2 (insulinoma-associated antigen 2), insulin and steroid 21-hydroxylase (21-OH) by radioimmunoassay (RIA), to thyroglobulin (Tg) and thyroperoxidase (TPO) and anti-transgutaminase (TRG) by chemiluminescence method, to parietal cells (PCA), adrenal cortex (ACA) and islet cell by indirect immunofluorescence (IFL). Anti-platelets Abs were tested by enzyme-linked immunosorbent assay (ELISA). The control group was made up of 314 healthy blood donors recruited from the Blood Transfusion Centre at OPBG; they had no history of autoimmunity and no autoantibodies were detected in their serum. All control subjects were matched for sex, age, ethnic origin and geographical area. All enrolled patients and controls in this study were unrelated. Informed consent was obtained from all those who took part in the present investigation before sampling.
2.2. Genotyping for rs30187 ERAP1 Polymorphism
Peripheral blood was collected in EDTA tubes. Genomic leukocyte DNA was extracted from whole blood of patients and controls by QIAamp DNA blood mini kit (Qiagen, Hilden Germany). SNP rs30187 was genotyped by using Taqman® SNP genotyping assays (assay ID C_3056885_10, Applied Biosystems, New Jersey, USA) according to manufacturers protocols. PCR reactions were carried out in a total volume of 20 μl containing 10 ng of genomic DNA, 1X TaqMan Universal PCR Master Mix No Amp Erase UNG and 0.625 μl Taqman® SNP genotyping Assays primers 40× and FAM/VIC labeled probes by Applied Biosystems as Assays-by-Design™ (Applied Biosystems). Genotyping reactions were performed with an AB 7900 HT, and the allele call by the analysis of allelic discrimination plots with AB SDS 2.4 software (Applied Biosystems, Foster City CA94404). All assays were performed in 96-well plates including replicate genotype known and negative control samples.
2.3. Statistical Analysis
SNP genotype data were assessed by Chi-squared (χ2) test and χ2 test with Yates’ correction (genotype test and allele test). Results were analyzed using the GraphPad Prism software version number 5 (San Diego, California, USA). A result with p < 0.05 was considered statistically significant.
3. RESULTS
The study population was 234 patients and 314 healthy controls. The mean actual age of T1D patients was 16.8 years (ranging from 2.6 to 39.1 years). Most of the patients were recruited at disease onset with a mean age at disease onset of 9 years. The mean age of the control group was 25 years (ranging from 18 to 39 years). Demographic and clinical characteristics of patients are shown in Table 1.
The T1D cohort population included 118 males and 116 females. In the female group of 116 patients, 82 patients were recruited at the disease onset, while 34 had long-standing disease. In the male group of 118 patients, 86 were selected at the disease onset, while 32 had long-term disease. T1D patients presented associated autoimmune manifestations (Table 1). Autoimmune gastritis was confirmed by the presence of PCA Abs in 2 female patients. In addition to T1D 46 patients were affected by autoimmune thyroid diseases (autoimmune polyglandular syndrome Type III variant): two patients were affected by Basedow’s disease and 44 patients were affected by Hashimoto’s thyroiditis confirmed by the presence of circulating Tg and TPO Abs and echografic pattern of diffuse hypoechogenicity. In association with T1D, 21 patients developed celiac disease confirmed by the presence of TRG Abs. 4 female patients manifested vitiligo. In addition to T1D one patient was affected by ulcerative colitis, one by autoimmune leucopenia confirmed by the presence of anti-leukocyte abs and one patients was affected by autoimmune thrombocytopenia confirmed by the presence of anti-platelets Abs. The genotype and allele distribution of rs30187 ERAP1 SNP in T1D patients compared to controls are shown in Table 2. The SNP did not significantly deviate from HardyWeinberg equilibrium. We detected rs30187 SNP G/G homozygotes in 90 patients (38.4%) and 114 controls (36.3%); rs30187 SNP A/G heterozygotes in 101 patients (43.1%) and 155 controls (49.3%); rs30187 SNP A/A homozygotes in 43 patients (18.37%) and 45 controls (14.3%). These differences in frequency distribution were found to be not statistically significant. Similarly no significant association was noted in allele frequencies of rs30187 SNP among T1D patients and control (Table 2) being the alleles equally distributed among patients and controls (the allele A was detected in 187 patients (40%) and 245 controls (39%), whereas the allele G was detected in 281 patients (60%) and 383 control (39%). These results suggest that ERAP1 has a minor contribution in the TD1 pathogenesis.
4. DISCUSSION
Fung et al. [22] analysed 17 SNPs from 16 gene re-
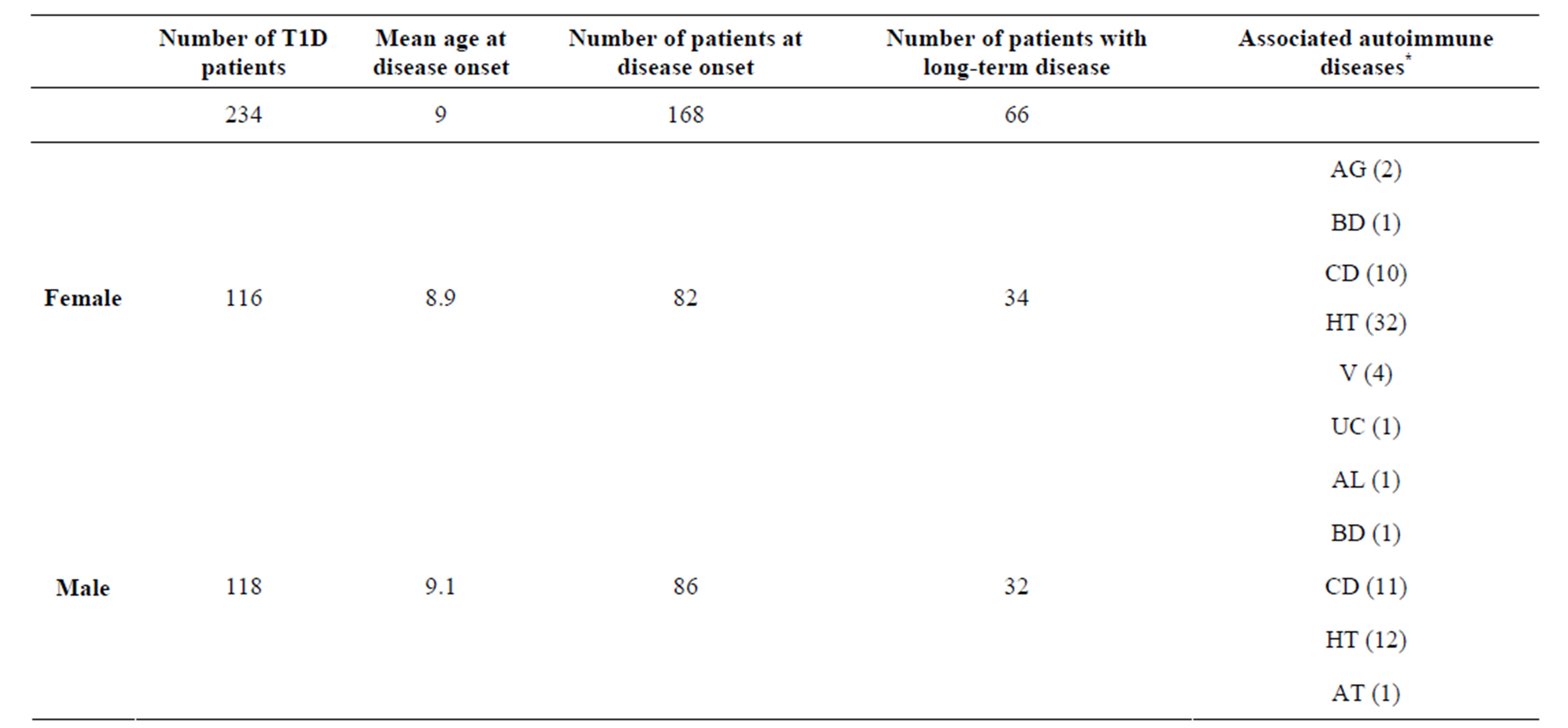
Table 1. Clinical characteristics of T1D patients.
*AG: autoimmune gastritis, BD: Basedow’s disease, CD: Celiac disease, HT: Hashimoto’s thyroiditis, V: Vitiligo, UC: Ulcerative colitis, AL: Autoimmune leucopenia, AT: Autoimmune thrombocytopenia.

Table 2. The distribution of genotype and allele frequencies of the rs30187 SNP of ERAP1 among T1D patients and controls.
aChi-square test; bChi-square test with Yates’ correction.
gions that were previously considered to be related to the pathogenesis of several autoimmune conditions including AS, rheumatoid arthritis (RA), systemic lupus erithema tosus (SLE), MS, and celiac disease to determine whether they could be associated also with T1D, since there was previous evidence of overlapping risk alleles across autoimmune diseases. Analysed genes included: ERAP1 (rs30187), found associated with AS (vide supra); CD58 (rs12044852), found associated with MS; STAT3 (signal transducer and activator of transcription 3, rs3816769), found associated with CD, IL-1A (rs17561) found associated with AS, MMEL1-TNFRSF14 (membrane metallo-endopeptidase-like 1 tumor necrosis factor receptor superfamily, member 14 herpesvirus entry mediator) (rs3890745), STAT4 (rs7574865) and TNFAIP3 (rs6920220 and rs10499194), which have been associated with both RA and SLE [15]. Among the others, the 7q23 gene desert (rs11761231), CCL21 (rs2812378), TRAF1/C5 (rs3761847), CDK6 (rs42041), KIF5A (rs1678542) and CD40 (rs4810485) found associated with RA were considered. In addition BANK1 (rs10516487), FAM167A (rs13277113) and ITGAM (rs9888739) found associated with SLE were also included in the investigation [22].
8010 cases and 9733 controls were analysed [22]. All cases (from the Juvenile Diabetes Research foundation/Welcome Trust Diabetes and Inflammation Laboratory’s British case collection) and controls were of self-reported white ethnicity and chosen to be matched geographically. Some evidence for an association with T1D was detected in the regions containing the following genes: 2q32/STAT4, 17q21/STAT3, 6q23/TNFAIP3 and 12q13/KIF5A/PIP4K2C. Evidence for some association with T1D was found in the region containing 5q15/ ERAP1 (rs30187, ARTS1). ERAP1 was also tested for interaction with HLA class I, especially with HLA-A and HLA-B, but no evidence for an interaction was observed by the authors [22].
In our observational study conducted in 234 patients and 314 controls of Italian ancestry, we could not replicate the association of the intergenic ERAP1 SNP rs30187 found by Fung et al. (2009) [22], suggesting a minor contribution of this gene to the pathogenesis of T1D in the Italian population. Failure of our study to find a definitive association could be due to insufficient power within the sample. To define the putative association of the ERAP1 polymorphism with T1D development large population-based case-control studies, necessitating follow-up from different ethnicities, will be required. On a speculative basis other polymorphisms within the ERAP1 gene may be involved in the susceptibility to T1D.
The relevance of the putative pathogenetic role of proteolytic pathways such as ERAPs in autoimmunity remains to be ascertained. In particular the role of SNPs on ERAPs that have been linked with predisposition to autoimmune disease has to be unravelled. How ERAP1/2 SNPs affect disease pathogenesis and which is in addition the diagnostic and prognostic value of their detection is at present unknown. In particular, future investigations are required to study their effects on protein function and antigen processing.
Furthermore cross-talk among different proteolytic pathways was recently highlighted i.e. components processed in the ubiquitin/proteasome system possibly engaged in autophagic pathways [23]. Recent data, in particular, emphasized the putative role of autophagy in the pathogenesis of autoimmune diseases [24] since the already reported discovery of polymorphisms in autophagy-related genes in patients affected by disorders such as inflammatory bowel diseases (IBD), RA, psoriasis, vitiligo, MS and SLE [25]. Recent data also demonstrated the requirement of autophagy in thymic epithelial cells (TECs) for loading endogenous antigens onto MHC class II and that this mechanism was critical for negative selection of CD4 T cells [26].
Deregulated autophagy was detected in T cells from lupus-prone mice and also found in T cells from patients affected by SLE possibly contributing to the survival of autoreactive T cells during lupus development as also shown in the experimental autoimmune encephalomyelitis model [27,28].
These observations open perspectives in unraveling the pathogenesis of autoimmune diseases through the combined analysis in GWAS of genes related to different proteolytic pathways.
5. CONCLUSION
We could not replicate in T1D patients from Continental Italy, the association of the ERAP1 polymorphism rs30187 previously reported by Fung and colleagues [22]. Our results suggest a minor contribution of the polymorphism to the pathogenesis of the disorder.
6. ACKNOWLEDGEMENTS
This study was supported by the Italian Ministry of Health. We acknowledge the Blood Transfusion Centre at OPBG for providing samples from blood donors.