Determining the specific status of the Iberian sturgeons by means genetic analyses of old specimens ()
1. INTRODUCTION
The identification of sturgeon species inhabiting a certain geographical region has interest not only from the basic scientific standpoint but also for the conservation and recovery of this group of ancient fish so important from the evolutionary as well as the economic perspective [1]. Thus, the specific status of the Iberian Peninsula sturgeons is a debatable matter because, bearing in mind that they are currently almost extinct, it becomes necessary to analyse old museum specimens and even archaeological remains. In this sense, during the second half of the 20th century, it was traditionally considered that in the seas and southern rivers of Western Europe and, more concretely, in the southern Iberian Peninsula, there was only one sturgeon species, Acipenser sturio (Linnaeus 1758). However, from end of last century, the idea arose that until recently at least two species could have coexisted. In fact, based on morphologic and mainly genetic studies (including mitochondrial and nuclear markers) of old museum specimens of sturgeons from this region, it has been shown [2-4] that, in addition to specimens belonging to A. sturio, it is possible to find specimens belonging to another species, A. naccarii (Bonaparte 1836). This situation had been previously proposed by different authors who historically, although forgotten, cited A. naccarii in the Iberian Peninsula [5-13]. All these results would indicate that, in recent historical times, this latter species (A. naccarii), until now considered only endemic to the Adriatic region, would also have lived in rivers of the Iberian Peninsula.
However, these results have been questioned partly by other studies, which have not provided data to indicate the presence of the species A. naccarii in this region [14-16]. Finally, recently Ludwig et al. [17] studying the mitochondrial region control in five scutes of sturgeons from archaeological locations of historical times in the Iberian Peninsula, have recently found only mitochondrial haplotypes of A. sturio. Therefore, it becomes necessary to continue delving into the analysis of this issue.
In this work, our group, which has contributed to opening this new vision of the distribution of sturgeons in Southern Europe (i.e. the coexistence of A.naccarii with A. sturio), analyses and discusses the attempts to obtain eight molecular markers (mitochondrial and nuclear) in seven old specimens of historic and prehistoric times in southern Spain. These molecular markers are compared in several sturgeon species. Thus, the results previously reported by our group have been corroborated in four historical specimens. In addition, we have tried to clarify the specific status of three new sturgeon samples from archaeological sites. Emphasis is placed mainly on the positive results for one of these samples, in a scute of a very old specimen dating from 1500 BC, which again verifies the presence of the species A. naccarii in this region.
2. MATERIALS AND METHODS
2.1. Samples
In this work, DNA was extracted from seven old sturgeon specimens from the southern Iberian Peninsula. Four of the specimens analysed had been captured in the Guadalquivir River, EBD8173, EBD8401, EBD8174 and UGP. Three of them (labelled EBD), captured in the 1970-80s, are conserved in the Biological Station of Doñana (Spain). The samples EBD8173 and EBD8401 are preserved in ethanol, whereas the EBD8174 is a dry skin. The fourth sample (labelled UGP) is a skin conserved in the Department of Biology Animal of the University of Granada and was also captured in the Guadalquivir River (19th century).
Additionally, three prehistoric samples are analysed for first time. One of them corresponds to a scute from 1500 BC which was found in the Acinipo archaeological deposit (Ronda, Malaga, Spain) (Figure 1). The archaeological deposit of Ronda la Vieja (called Acinipo, the name of the Roman city built on this site; [18]) is located in the depression of Ronda, 20 km from the city. The site

Figure 1. Scute dated in 1500 BC found in the archaeological deposit of Acinipo (Ronda, Malaga, Spain).
is situated on a large limestone plateau, which provides a strategic view of the surrounding territory and provides communication with other areas, including the countryside of the Guadalquivir River. The bony sample of sturgeon analysed corresponds to the archaeological phase Acinipo III [19], prior to the Phoenician colonization around the second half of the II millennium B.C. Although it is difficult to assign its origin to the Guadalquivir River, the dates and the zone where it has been found would indicate its origin from this river. Finally, an attempt was made to extract DNA and amplify the different molecular markers from two very old scutes of sturgeons found at another prehistoric deposit (Cave of Nerja, Malaga, Spain). The Cave of Nerja has a long ichthyoarchaeological record of the excavations made basically in the room of the Vestíbulo [20] on the stratum VII (about 12,000 years old). This level is correlated with Magdalenian occupation in the cave [21].
2.2. DNA Extraction
The extraction and purification of DNA was carried out using ancient DNA techniques and according to the protocol described in Martínez-Espín et al. [22]. The first step consisted of cleaning the tissue samples in a polymethacrylate (PMMA) box. A miniature Dremel drill was used to eliminate any polluting agents adhering to the surface. Then, the tissue samples were pulverized in liquid nitrogen using a Freezer Mill. After pulverization, the powdered sample was transferred to a sterile 15 ml conical polypropylene tube.
To improve DNA recovery, in older samples (the scute from Acinipo and the two scutes from Nerja), we made some changes in the protocols. For these three samples, a protocol was adapted for demineralization of skeletal remains frequently used in mommies and historical identification [23,24]. To minimize the possibility of contamination by contemporary DNA of extraneous sources, these samples were extracted in the minimal-humanremains laboratory, where an animal sample had never before been extracted. Here, possible contamination was eliminated from the old samples. Only one specimen was cleaned and processed at the same time and a negative control was included with the analysis of each specimen. After adding demineralization buffer, the samples were incubated on an orbital shaker at 56ºC for 20-30h. The tubes were angled during agitation to ensure thorough mixing. At the beginning of the extraction, we first added 50 µl of proteinase K (20 mg/ml) and 25 µl again 18 h later. The extracts were purified using sterile water washes in Microcon YM-30 Millipore centrifugal filter units; in the other samples, Microcon YM-100 was used. As a final point, the concentrator was discarded, and 200 µl of the purified DNA were obtained. In this case, many inhibitors were also obtained owing to the fact that tissue is adsorbed into a mineral matrix, after the death of the animal. The following step was the purification with the GENECLEAN® (BIO 101) for Ancient DNA Kit (using the recommended protocol). To guarantee the absence of inhibitor, the Quantifiler® kit for 7500 Real-Time PCR (Applied Biosystems) was used. The Internal Positive Control detectors indicated the absence of PCR inhibitor in all samples.
2.3. Amplification, Cloning, and Sequencing of Molecular Markers
For each specimen an effort was made to characterize the following genetic markers: 1) four nuclear markers corresponding to two satellite-DNA families: the family HindIII [25] and the family PstI [26]; non-transcribed sequences of 5S ribosomal gene (NTS) [27] and 230 base pairs from nuclear DNA flanking the microsatellite Aox-23 [28]. 2) four mitochondrial markers corresponding to two fragments of the cytochrome b gene of 212 bp and 265 bp, respectively [29,30], one fragment of 210 bp corresponding to the mitochondrial region control, d-loop, [30], and one fragment of the 12S ribosomal gene of 139 bp [16]. In each case, the PCR reactions were carried out with the amplification conditions described in each of the references.
Each marker was cloned using the vector TOPO TA (TOPO TA Cloning® kit PCR® 2.1) and were used to transform the cells DH5α of E. coli, according to the supplier recommendations (Invitrogen Carlsbad, CA, USA).
Recombinant plasmids were sequenced on both strands using Big DyeÒ Terminator Cycle Sequencing Kit (Applied Biosystems) and T7 and M13 primers in an ABI PrimsÒ 3100-Avant Genetic Analyzer DNA Sequencer (Applied Biosystems).
2.4. Sequence Analysis
Multiples alignments of sequences obtained from the samples and reference sequences from GenBank database were performed using ClustalX software [31]. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 4 [32]. Sequence divergences were calculated according to the Jukes-Cantor method and distance trees produced by UPGMA [33] and the neighbour-joining method [34].
3. RESULTS AND DISCUSSION
The (Table 1) presents a summary for all the seven sturgeon specimens from Iberian Peninsula analysed for different molecular markers. This table includes the data obtained in this new study, completed with the data from previous analyses made by us.
The specimen UGP (Table 1) had previously been analysed for four markers (HindIII and PstI satellite DNA family, 212-bp cytochrome b and 12S mitochondrial gene) and catalogued as A. sturio [4]. Considering nuclear markers, Garrido-Ramos et al. [4] analysed this specimen for the HindIII satellite DNA family, showing the lack of this repetitive sequence in its genome (its absence is characteristic of the species A. sturio; [35]) In the same study, these researchers showed that the sequences corresponding to PstI satellite DNA family analysed for this specimen UGP were grouped, in a phylogenetic tree, together with the sequences of A. sturio.
Now, nine clones have been sequenced for non-transcribed sequences of the 5S ribosomal nuclear genes (NTS), and their sequences were aligned with NTS ribosomal genes from other sturgeon species (Figure 2). Characteristic positions for A. sturio and A. oxyrinchus are present in the sequences isolated from UGP. Thus, in a phylogenetic tree based in genetics distances, all sequences belonging to this sample were grouped together with the NTS sequences of A. sturio (Figure 2).
Additionally, a new nuclear marker Aox23 locus [28] was amplified in this specimen. The sequence found, when compared with sequences of A. sturio and A. oxyrinchus taken from GenBank, proved similar to those of A. sturio (data not shown).
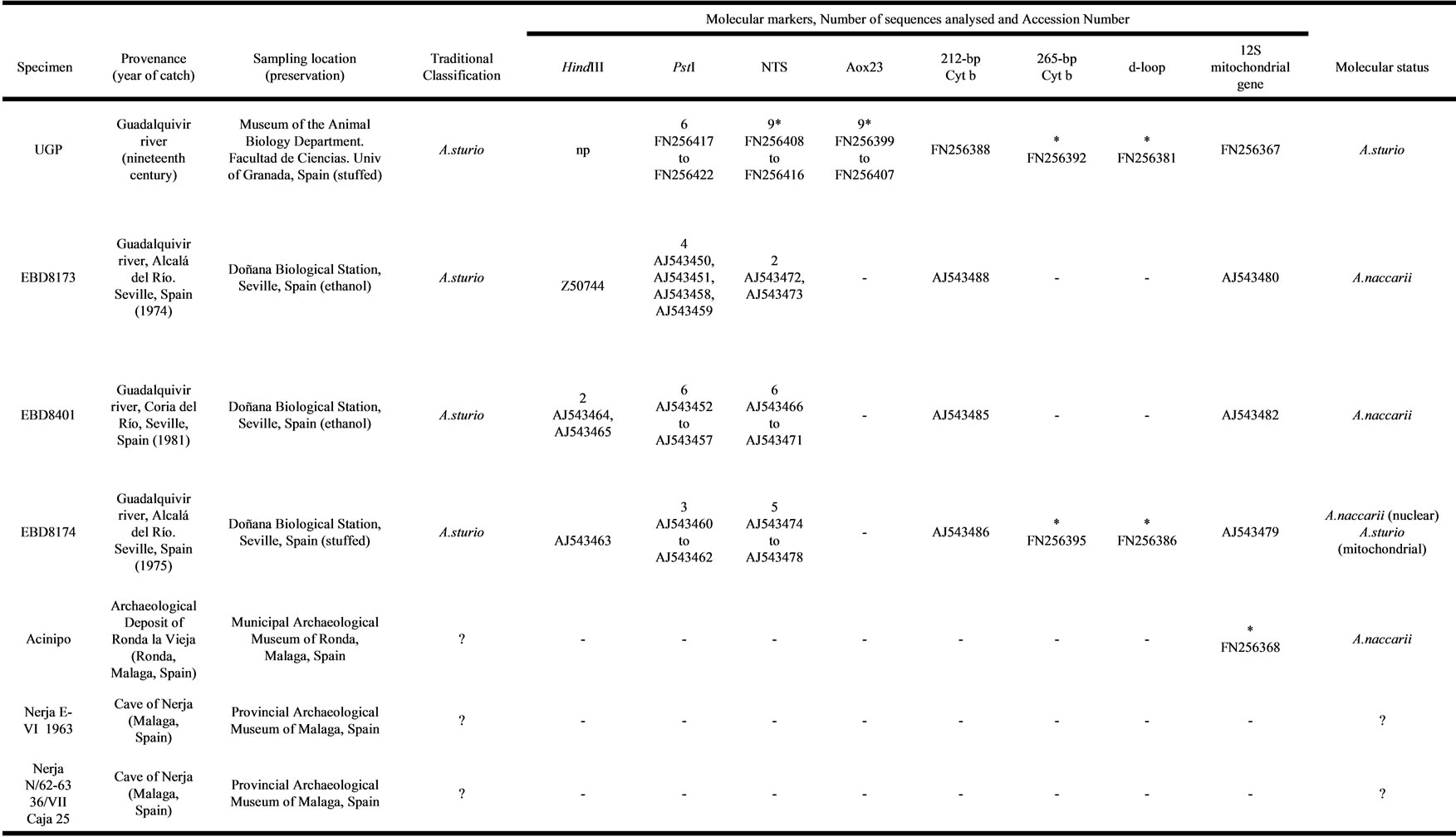
Table 1. Summary of sturgeon specimens analysed.
List of sturgeon specimens analysed, their current specific status and the results for the markers analysed in each specimen and the number of units sequenced (with their accession number) for each nuclear repetitive marker or the number of mitochondrial clones sequenced for each mitochondrial marker. np: not present; na: not amplified; the asterisk (*) shows the sequences found in this study; question mark (?) indicates unknown Traditional Classification and/or Molecular status.
With respect to mitochondrial markers, GarridoRamos et al. [4] analysed in this specimen the fragments of the mitochondrial DNA 212-bp cytochrome b and 12S gene and considered UGP as A. sturio. In the present work, two new mitochondrial markers have been amplified for this sample (265-bp cytochrome b and d-loop). It was found that all diagnostic positions for these markers correspond to the species A. sturio (Figures 3(a) and (b)).
Therefore, the results of eight nuclear and mitochondrial markers confirm the classification of this sample (UGP) as A. sturio. This affirmation is not surprising if we bear in mind, as mentioned in the Introduction, that the species A. sturio has been broadly described in most of rivers of the Iberian Peninsula.
On the other hand, previous molecular analyses carried out by our group in three samples from the Biological Station of Doñana (Table 1), identified two of them, EBD8173 and EBD8401, as A. naccarii, based both on the mitochondrial and on the nuclear markers [2-4]. Thus, the samples EBD8173 and EBD8401 have the HindIII satellite DNA family in their genome. This satellite DNA, as commented above, is absent in the A. sturio genome [2].
The presence of this repetitive sequence means that these two samples cannot be assigned to A. sturio, the only species that had previously been considered to live in the rivers of the Iberian Peninsula. Additional results using the markers PstI satellite DNA, non-transcribed sequences of 5S ribosomal gene (Figure 2), 212-bp cytochrome b and 12S mitochondrial gene (Figures 3(a) and 4) confirmed that EBD8173 and EBD8401 belong to A. naccarii [3,4].
However, the sample EBD8174 (Table 1) is a special specimen from the genetic perspective. For all the nuclear markers analysed to date, this sample EBD8174 cannot be assigned to A. sturio but to A. naccarii. The presence of HindIII satellite DNA family and the fact that all the sequences corresponding to the nuclear markers (the HindIII itself and satellite PstI and NTS) are not grouped with A. sturio but with A. naccarii are indicative of this fact [3,4]. However, previous mitochondrial DNA studies using 212-bp cytochrome b and 12S mitochondrial gene DNA markers [3,15,16], conclude that, in this specimen, mitochondrial DNA markers are similar to A. sturio.
Thus, the results of nuclear and mitochondrial DNA are contradictory in this specimen because, the nuclear

Figure 2. UPGMA tree based on NTS sequences of 5S ribosomal nuclear genes.
UPGMA tree based on NTS sequences and Juckes Cantor distances calculated in MEGA 4. The tree shows the close relationships between the 9 NTS sequences from UGP (▲) specimen and NTS sequences of A. sturio (STU AJ550044) and A. oxyrinchus (OXY AJ555397), and between the 13 NTS sequences from the three EBD specimens -EBD8173 (■), EBD8174 (●) and EBD8401 (♦)- and NTS sequences of A. naccarii (NAC AJ550039) and other sturgeon species as A. transmontanus (TRA AJ555360), A. baerii (BAE AJ555351), A. gueldenstaedtii (GUE AJ555353), A. stellatus (STE AJ555385), Huso huso (HUS AJ555358) and A. ruthenus (RUT AJ555393). The code name species and the accession number are show into parenthesis. Numbers indicate bootstrap support for each node (10000 replicates).
DNA markers indicate its assignment to A. naccarii but mitochondrial DNA markers show identities to A. sturio. To confirm this situation, we analysed two new mitochondrial markers, 265-bp cytochrome b and d-loop (Figures 3(a) and (b)). And the results coincided with previous ones, demonstrating that this sample corresponds to A. sturio for all mitochondrial markers. In fact, in the mitochondrial sequences analysed in this study (265-bp cytochrome b and d-loop), we found positions fixed with those of the species A. sturio Thus, the specimen EBD8174 could be considered a “mosaic” sturgeon: having nuclear characteristics of A. naccarii but mitochondrial markers of A. sturio. Hybridization or introgression processes between A. sturio and A. naccarii could explain this phenomenon. In sturgeons, genetic evidence of hybridisation phenomena between sympatric sturgeon species has been shown for example in Arefjev [36], and more recently between A. ruthenus and A. baerii in the Danube River [37].
Also, similar introgression processes have been described previously in the Adriatic region (A. gueldenstaedtii introgressed into the A. naccarii) [38] and in the population of the Baltic Sea of A. sturio (A. oxyrinchus introgressed into the A. sturio; [39]).
Finally, we have tried to clarify the specific status of three samples from archaeological sites (Table 1). Two of these samples (about 12,000 years old found at an older prehistoric settlement, the Cave of Nerja) were not successfully analysed. Unfortunately, none of the markers used could be characterized for these samples. However, we succeeded in amplifying a fragment of the 12S mitochondrial gene from the prehistoric scute (Ronda, Malaga, of about 3500 years of antiquity) found at the archaeological site of Acinipo (Table 1). These results are tentative because the first samples were very old and it was difficult to extract enough quality DNA to amplify the molecular markers. However, in previous studies some samples with similar antiquity at Acinipo, have been used successfully in species identification [17,40].
The 12S mitochondrial gene obtained from the Acinipo sample was compared with other 12S sequences from different species of sturgeons in the GeneBank database. The diagnostic positions for this marker did not coincide with A. sturio, ruling out its assignment to this species (Figure 4). In fact, all diagnostic sites coincided with A. naccarii, although they are not exclusive of this species, sharing them with other sturgeon species such as A. gueldenstaedtii, A. baerii, A. persicus and A. nudiventris with a distribution far away from the Iberian Peninsula. Thus, in a phylogenetic tree, based on genetic distances, the sequence from the Acinipo scute is grouped with the sequences from A. naccarii (Figure 5).
4. CONCLUSIONS
The nuclear and mitochondrial markers show that the specimens EBD8173 and EBD8401 belong to the species A. naccarii, and the sample UGP to A. sturio. The specimen EBD8174, using mitochondrial markers can be catalogued as A. sturio, or as A. naccarii according to nuclear markers. Hybridization or introgression processes between A. sturio and A. naccarii, could explain this phenomenon, common in sturgeons in these species. On the other hand, we were able to analyse the 12S mitochondrial marker for the ACINIPO sample (3500 years old) demonstrating that it belongs to species A. naccarii. These analyses provide insights into the existence of specimens belonging to A. naccarii in the southern Iberian Peninsula in historic (EBDs samples) and prehistoric (ACINIPO) times. Thus, our analyses confirm old references mentioning the presence of A. naccarii in the Iberian Peninsula [5-13]. Therefore, although A. naccarii is currently considered endemic of the Adriatic Sea, in the past it could have had a broader distribution area, extending to the Iberian Peninsula, including the Gua


Figure 3. (a) Alignment of sequences of a 265-bp cytochrome b fragment; (b) Alignment of partial d-loop sequences.

Figure 4. Alignment of sequences of 12S mitochondrial gene from Acinipo.
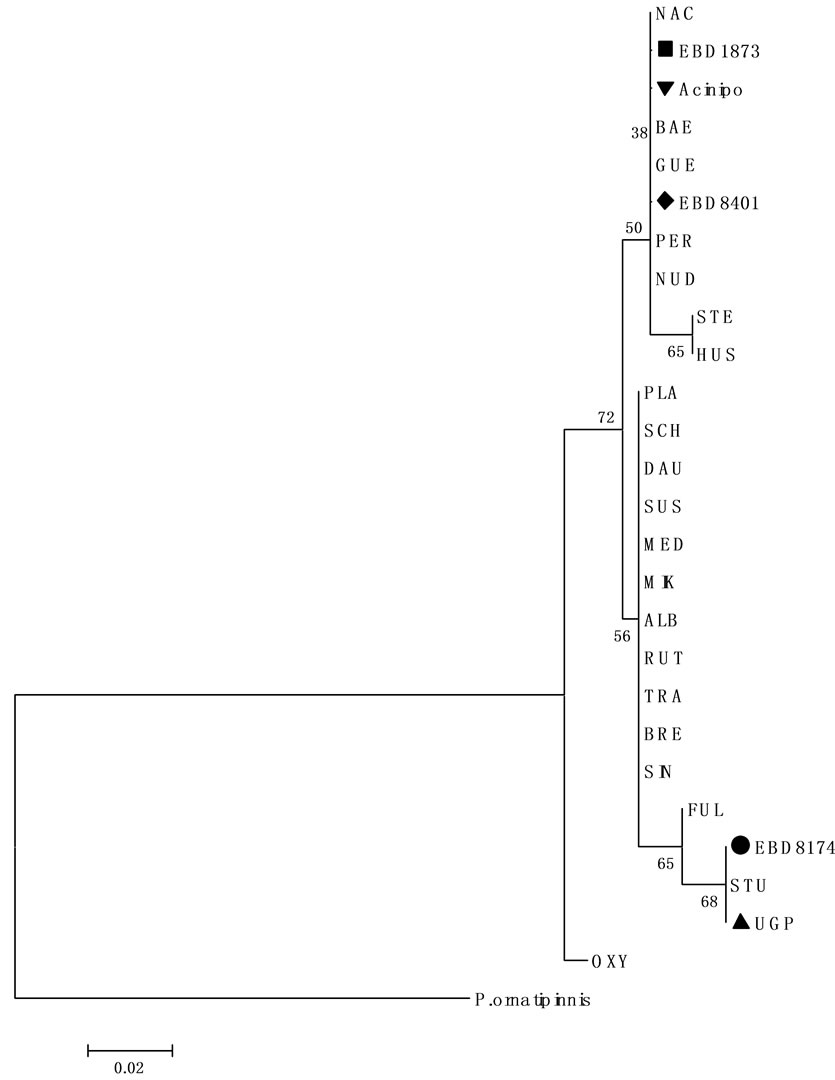
Figure 5. Neighbour-joining tree based on 12S mitochondrial gene sequences.
Neighbour-joining tree based on 12S mitochondrial gene sequences and Jukes-Cantor distances calculated in MEGA 4. The tree shows the close relationships between sequences from Acinipo (▼), EBD8173 and EBD8104 with A. naccarii and between sequences from UGP and EBD8174 with A. sturio. Numbers indicate the bootstrap support for each node (10000 replicates). Polypterus. ornatinnis (Bichir NC001778) is used as outgroup.
dalquivir River. Similarly, A. sturio was distributed not so long ago throughout Europe whereas, at the present, only one population exists, in the Gironde-GaronneDordogne River, France [41-44]. Furthermore, to propose a broad distribution area for A. naccarii is consistent with the general observation that most sturgeon species inhabited vast areas of continents and river basins [45]. Thus, observations based on molecular analyses, as we present in this paper, or the finding of an “American” species in Europe (i.e. the movement of A. oxyrinchus into Europe during the Little Middle Ages [46,47]), require more studies in order to establish a more complete vision of the distribution of different sturgeon species in Western Europe.
5. ACKNOWLEDGEMENTS
This research has been financed by grants of the Junta de Andalucía, Consejería de Innovación, Ciencia y Empresa (Proyecto de Investigación de Excelencia P07-CVI-03296) (F. Robles is a postdoctoral grant holder in this Project). Nerja Cave was analysed in the Project “Revisión, estudio y contextualización cronoestratigráfica de los restos arqueológicos procedentes de las antiguas excavaciones del Patronato de la Cueva de Nerja”, authorized by Consejería de Cultura de la Junta de Andalucía to one of the authors (MCS). We also thank our colleague D. Nesbitt for revising our English text.