Probabilistic Modelling of COVID-19 Dynamic in the Context of Madagascar ()
1. Introduction
Since its emergence in Wuhan, coronavirus 2 (SARS-CoV-2) has shaken up scientific communities around the world [1]. Currently, there are more than 100,000 articles published according to the database “COVID-19 primer” (https://covid19primer.com) most of whom are interested in modeling and forecasting [2] [3] [4]. The case of less developed countries in sub-Saharan Africa such as Madagascar is still less studied because of insufficient data. For Madagascar, since the first case in March 2019, the COVID-19 has affected more than 14,696 people until 30 August 2020 according to official statistics. Of the contaminated, 13,709 are cured with 187 deaths officially declared by the ministry. The cure rate is, therefore, close to 93.28% and the mortality rate is about 1.30%.
This statistic is widely questionable given the insufficient number of tests on the big island. The absence of a massive test and its management principle make the choice of the procedure for responding to the disease quite particular in Madagascar. Socio-economic factors do not allow containment measures to be strictly applied. This specificity extends to the modeling approach of the evolution of the pandemic to represent reality [5] [6]. Studies have been conducted on the identification of the various symptoms and clinical signs of COVID-19 to anticipate the detection of confirmed cases.
From the compartmental models of epidemics introduced by Daniel Bernoulli [7]; several extensions have been proposed to explain the evolution of more diseases [8] [9] [10]. These models have been widely used by scientists around the world to model the evolution of the COVID-19 epidemic. Several compartments can be used depending on the modeling context. Faïçal et al. [8] consider an 8 compartments model and Kyrychko et al. arrived up to 10 compartments [11]. The number of parameters as well as the complexity of the model increase with this number of compartments. Our challenge in this article is to have a relatively simple, understandable but realistic model in the context of Madagascar.
As in all epidemic models, the basic reproduction number gives us information on the spread of the disease. In order to have a better estimate of this parameter, we combine several approaches starting from a deterministic model in order to study its stochastic extension. For the deterministic case, the main results on the ordinary differential equation make it possible to identify important properties on the propagation of the disease [12] [13]. The basic reproduction number is considered to be constant.
Considering the fact that people do not have the same sensitivities to contact with the virus and different factors can influence the rate of spread of the disease; the use of probabilistic models seems realistic to us. From section 4, we focus on the stochastic extension of the deterministic model, with the introduction of random noise. The techniques used are detailed in the article by Alen et al. [14].
In [15], with its six-compartment model, authors introduced a similar approach to model the evolution of the new coronavirus in the island of Mayotte (French). We have adapted this model for the case of Madagascar with our seven compartments epidemic model. We propose new methods for simulation and estimation of parameters. The models will be inferred with official data on COVID-19 in Madagascar, during the critical moment of the epidemic, i.e. between March and August 2020.
2. Preliminaries
2.1. Local Context of the COVID-19 Epidemic in Madagascar
Madagascar remains vulnerable to coronavirus attack. Despite the small number of cases detected, the impact of the new coronavirus on the Malagasy health system is considerable. The country is still at risk of an unmanageable and dangerous health crisis.
As soon as the virus appeared in Madagascar, the CCO took the initiative to introduce restrictions in order to limit the spread of the virus. Studies have shown that this initiative is early and has not provided more information on how the virus spreads.
According to official data, during the first wave, between March and August 2020, the outbreaks are in two regions: Analamanga region (Antananarivo, capital) and Antsinana region (Tamatave) (Figure 1).
Much of the data used in this article comes from these two strategic regions of Madagascar: Antananarivo the capital and Toamasina the economic capital. Figure 2 and Figure 3 represent the evolution of new cases observed in these two regions.
These graphs show that the epidemic has not had the same dynamic in these two major regions of Madagascar. It first hit the east side of the big island before entering the highlands.
For the entire territory of Madagascar, the SRAS-COV2 coronavirus epidemic has experienced several phases. The numbers of new cases and deaths were considered constant during the first trimester. These numbers followed an upward phase from June 2020. The impact of the epidemic was major in July 2020. The peak of this first wave is assumed to have reached around the beginning of August 2020, see Figure 4.
Figure 5 describes the cumulative data observed for all 22 regions of Madagascar.
2.2. Modelling approach
The effectiveness of SIR (Susceptible-infected-removed) models has already been shown in epidemiological modelling problems. Several modifications have been proposed based on hypotheses on how the disease is transmitted [7] [8].
To limit the spread of the COVID-19 virus, Madagascar’s strategy is to isolate all suspected cases and hospitalize confirmed case. Given the inadequacy or lack of mass screening (PCR test), the effects of this strategy remain questionable. Many people with the virus are asymptomatic and show no signs of infection.
In line with this strategy, we propose in this article a model with seven compartments:
• S (Susceptibles): individuals who are not infected but at some risk of becoming infected;
• E (Exposed): individuals who have been in contact with the virus but not yet infectious until after a latency period;
![]()
Figure 1. Official data observed for the 22 regions of Madagascar during the first wave of the covid epidemic 19.
![]()
Figure 2. Evolution of new cases of COVID-19 in the Antsinanana (Toamasina) region during the first wave.
![]()
Figure 3. Evolution of new cases of COVID-19 in the Analamanga (Antananarivo) region during the first wave.
![]()
Figure 4. Daily evolution of the numbers of new cases, healed and dead.
![]()
Figure 5. Cumulative numbers of confirmed, cured and dead cases for all 22 regions of Madagascar during the first wave.
• I (Infectious): individuals who can infect and are not yet hospitalized or isolated,
• H (Hospitalized): confirmed and hospitalized (isolated) cases;
• A (Asymptomatic): infected individuals but do not have symptoms (not isolated);
• C (Cured): healed confirmed cases;
• D (Death): deaths due to COVID-19.
Diagram in Figure 6 describes the evolution of our system according to the flows between the different compartments. The particularity of this model among others are first of all the separation of healthy (susceptible) individuals and individuals who are not yet sick but already in contact with the virus (Exposed). We have also put in another compartment infected individuals asymptomatic but who can transmit the virus.
Using the assumptions of Manou-Abi and Balicchi in [15] all the models’ parameters is summarized in Table 1.
The basic reproductions number R0 corresponding to the model will be estimated using the officiad data recenced by the CCO-COVID-19. It represents the average number of secondary cases produced by a fundamental case over its infectious period. It is the initial growth rate of the epidemic.
3. Classic Deterministic Model
3.1. Deterministic SEIR Modified Model
As a reference, we will consider the deterministic case adapted to the context of.
![]()
Figure 6. COVID-19 modified SEIR model with seven compartements.
![]()
Table 1. Parameters of the COVID-19 SEIR modified model.
Madagascar. This type of model is widely used in mathematical modeling of the spread of the COVID-19 [5] [16].
At a moment t, let
et
the number of individuals in the compartments “Susceptible”, “Exposed”, “Infections”, “Asymptomatic”, “Hospitalized”, “Cured” et “Death”.
Assume that the total number of population is constant, i.e.,
, (1)
with
, for any momentt.
In a time interval
, the evolution of the system is described by Equation (2):
(2)
with:
-
the number of confirmed cases (isolated, hospitalized),
-
the number of asymptomatic cases,
-
the number of new infections in the interval
.
By grouping asymptomatic cases and healed people into R(t) (“removed”):
(3)
the simulation of the model with an extract from the population, in an environment where the virus circulates, gives the graphs of Figure 7.
3.2. Estimation of the Parameters and Simulation
To estimate the infection rate β and recovery rate γ in the SIR sub-process, we use the method introduced by Kermarck and McKendick in [17] using the observed data.
![]()
(The parameters β and γ of the SIR sub-process).The number of basic reproductions R0 is obtained by
, (4)
after training the ODE system of Equation (3) with the data of the COVID-19 in Madagascar.
Note:If R0 < 1, the infected individual contaminates less than one other individual on average. This is why the epidemic is disappearing from the population.
If R0 > 1, then the virus continues to spread in the population.
For Madagascar, by running the model with the observed data and its population of about 26,026,000, we obtain:
et
, and
![]()
Figure 7. Simulation of the model with a population extract in an environment where the virus circulates.
The estimated number of basic reproductions with official data is quite small. This may be due to the specificity of the Malagasy on the vulnerability to the COVID-19. Figure 8 shows the simulation of the model with the epidemic parameters of Madagascar. Without intervention, the dynamics stabilize over the long term, see Figure 9.
4. Stochastic Model
In this section, we study the stochastic pattern of the propagation dynamics of COVID-19 in Madagascar with a continuous time Makov chain (CTMC) [18]. In complex and imprecise situations, the use of stochastic models is always a better alternative. These models are very useful in epidemiology [19] [20] [21].
4.1. Stochastic Evolution of the Spread of COVID-19
The spread and speeds of transmission of the new coronavirus vary widely from country to country. It depends on many factors such as demographic, socioeconomic, geographical, biologic and especially epidemic [22]. At the same time, individuals strive to find good ways to deal with the disease and adapt their behaviors to the different requirements of the riposte strategy.
We assume that factors external to the virus affect its rate of spread and turn it into a random variable. Thus, the process formed by transmission speeds
can be considered a stochastic process in continuous time with:
(5)
where B is a standard Brownian movement [23].
represents the number of infection transmitted by an infectious individual during a period dt. It is a normally distributed random variable [15]:
.
![]()
Figure 8. Simulation of the modified SEIR model Madagascar data and the estimated R0 = 1.2 parameter.
![]()
Figure 9. Long-term SEIAHD process dynamics.
4.2. Continuous-Time Markov Chain SEIR Modified Model
The works of Norris [18] and Durret [23] are good references to the theories on continuous time Markov chain. We use results that are demonstrated in these works.
For the SEIAHCD process of Figure 6, for each individual, consider the following events:
i) Exposition: a susceptible individual became exposed through contact with an infected individual, under a probability of p1
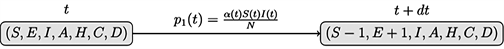
ii) Infection: an exposed individual became infected under a probability of p2
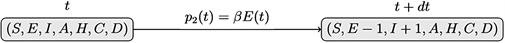
iii) Recovery: an infected individual has passed the incubation period and become an asymptomatic case, with a probability of p3
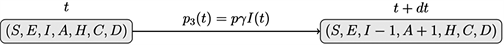
iv) Hospitalization: an infected individual is hospitalized with a probability of p4
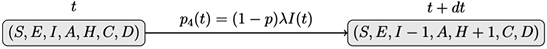
v) Healing: an individual hospitalized is cured of COVID-19
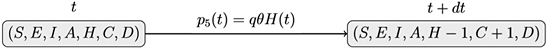
vi) Death: an individual hospitalized died of COVID-19
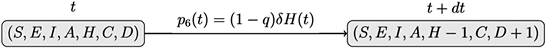
Events i), ii), iii), iv), v) et vi) correspond to jumps in processes
et
. Consider the following subset of events:
- Events after exposition:
, where
.
- Events after infections:
, where
, with
- Events after hospitalization:
.
In our modeling approach, we assume that events in each subset are independent uniformly distributed. Probabilities of jumps corresponding to the different events can be written:
- For G1:
,
,
.
- For G2
,
,
.
- For G3:
,
.
with the below assumption and with an additional hypotheses that the transition time are exponentially distributed, we can consider that processes
et
can be modeled by continuous time Markov chain. Algorithm 1 can simulate this model with the parameters estimated using official data. The result is in Figure 10.
5. Extending the Model to a Stochastic Differential Equation
5.1. Stochastic Equivalent of the Deterministic SEIR Model
The introduction of random effects into Equations (2) leads to a stochastic differential equation system. The deduced model is the stochastic equivalent of the modified SEIR model [24].
In a general way, a classic differential equation in explicit form
transforms into a stochastic differential equation by adding random noise.
This noise can be taken into account by a process of Winner Wt [25]:
, (6)
where
the average et
the covariance matrix.
![]()
Algorithm 1. Simulation of the continuous time Markov chain SEIAHCD model.
![]()
Figure 10. Simulation of a COVID-19 propagation model with Markov chains in continuous time.
1) First, consider the sub-model SEI (Susceptible, Exposed, Infected). Let
, associated random variables. For a small interval of time ∆t; transition probabilities are written:
(7)
We obtain:
(8)
where a Wienner i process.
2) For the sub-model “after infections”:
Note
the random variables associated with sub-model AH.
For a small interval ∆t. Transition probabilities are:
.
We obtain:
(9)
where W the independent processes of Winner.
3) For the last sub-model (compartments after hospitalization):
Note
random variables corresponding to the CD submodel. For a small inteval ∆t, the transition probabilities are:
The system becomes:
(10)
where W a Wienner process.
Figure 11 represents a simulation of the model through a numerical resolution of stochastic differential Equations (8)-(10).
5.2. Estimation of the Basic Reproduction Number R0
1) Estimate by the method of maximum likelihood:
In [26], the authors propose maximum likelihood method to estimate the number of basic reproductions of an epidemic. This technique is based on the assumption that the number of secondary cases caused by an index case is a Poisson distribution, with the expected value R0.
Let
the observation on consecutive time and ω generation time distribution. The number of basic reproductions R0 is estimated by a maximization of the log-likelihood:
(11)
where
.
The likelihood function must be calculated over a period of exponential growth, and the R-square deviance can be used to select the best period. For Madagascar, using the observed data, the maximum likelihood estimate (MLE) of the basic reproduction number is:
![]()
Figure 11. Simulation of the stochastic equivalent of the modifed model SEIR.
with 95% of confidence interval.
2) Bayesian estimate:
Introduced by Bettencourt and Ribeiro [27], this technique allows sequential estimate of the number of reproduction R. It is based on the assumption that the number of infections
is a discrete random variable and
is a Poisson random variable:
, where
the average duration of the infectious period.
With a non informative prior distribution of the basic number of reproduction R0, we have implemented an algorithm that can calculate the posterior distribution:
(12)
Knowledge about R0 is updated as new data is observed. On each new day, the posterior distribution after the previous day is considered to be the prior distribition. At any given moment, the Bayesian estimate can be calculated with the highest probability density interval.
As before, the method requires that the epidemic be in a period of exponential growth, i.e., it does not take into account the depletion of sensitivity. It implicitly uses an exponential distribution for generation time; and assumes a random mix in the population [28].
For our case, on the graph of Figure 12, the grayed area corresponds to the
![]()
Figure 12. Estimated number of temporal reproductions until 2020-08-22 with a Bayesian approach.
confidence interval and the black curve to the average number of reproductions in time function
. According to these results, the basic reproduction number in the Malagasy territory is lower than the references in neighboring countries [29] [30].
6. Conclusions
We have proposed a mathematical modelling framework for the spread of the COVID-19 epidemic in Madagascar taking into account local contexts. We developed a seven compartments model corresponding to the pandemic management strategies adopted by Malagasy authorities.
The implementation of containment and the imposition of various social distancing measures from the first outbreaks of the pandemic give a particular dynamic to the evolution of coronavirus in Madagascar. Due to insufficient screening tests, the detection rate of positive cases remains very low. There has been a sufficiently slow dynamic compared to other countries.
For the first deterministic model, the estimated basic reproduction number is close to 1.2. With the stochastic extensions of this model, we estimated values slightly below (near 1.1). These parameters suggest a low rate of spread of the epidemic during the first wave of coronavirus between March and August 2020 in Madagascar. According to official data, the epidemic appears to be under control. This deduction leaves much discussion given the lack of a massive test and the low rate of positive case detection.
Probabilistic tools provide a natural approach to modelling the evolution of the epidemic. Results already established in probability theory allow for more in-depth studies. The first stochastic model derived from a continuous-time Markov chain is reliable for a small-scale population. Managing transition time and events is a big challenge for a large number of populations. The stochastic differential equation (SDE) model is more interesting in this situation. Yet analyses of model characteristics, such as stability and convergence, are meriting further study.
For the proposed models, we were able to estimate the key parameters of epidemiological models such as the basic reproduction number. It can be interpreted as the measure of contagiousness and severity of the epidemic. Based on our results the Malagasy people are less vulnerable to the attack of the COVID-19. The epidemic is a little mild if we look at the official observed data.
With a Bayesian approach, we calculated the real-time estimation of the reproduction number. This time function of the
can be used to introduce control over the spread of the disease. This is also one of the perspectives of this work.