Taxonomy and Characterization of Fungi Isolated from Cocoa Beans during Fermentation in Saf’s Agroforestry System in the Amazon ()

1. Introduction
The northeast region occupies the first position with planted areas, with an estimated 440045.00 hectares of the cocoa planted area, but when it comes to productivity, the North region leads with 151163.00 tons, the state of Pará stands out in the current scenario, growing since 2000, taking the first position in 2021, Bahia superior with the production of 144216.00 tons, where Bahia occupies the second position with 106045.00 tons of cocoa [1] .
The growing cocoa industry in the state has become one of the most competitive in the world due to its high average productivity and low production costs. The North region leads the first position in 2021, superior Bahia state [1] . These factors are related to the preservation characteristics of cocoa production in the agroforestry system, manufacturing the cultivation of cocoa in Pará, an interesting alternative for sustainable rural development [2] .
Cocoa Agroforests are found in Asia, Latin America and Africa. Cocoa grows naturally in the lower strata of the rainforest with low light and high humidity. It is believed that the presence of shaded species in agroforestry systems filters access to light, softens the microclimate, and stores water and nutrients for the cocoa plant [3] .
To obtain a quality almond, cocoa processing is extremely important, and must be done well from harvesting, fermentation, drying and storage [4] .
The cocoa bean itself already contains microorganisms that act beneficially on the bean, as is the case of some fungal genera, their participation occurs in the first 48 hours of the fermentation process, among them are the yeasts and filamentous fungi that grow in acidic pH [5] .
The growing increase in the consumption of agricultural products and the demand for quality products is intensifying more and more. One of the factors that interfere with good post-harvest results is the presence of pathogenic microorganisms, which is often present in the product’s maturation process and perpetuates until the end of its processing.
In some cases, fungi can produce mycotoxins and aflatoxins, which are fungus-generating products with toxic and harmful effects on health. Aflatoxins occur naturally in nuts, cereals and rice under conditions of humidity and high temperatures, especially during the storage process. This mycotoxin is harmful and has the ability to cause liver disease [5] .
The microbiota is the established microorganisms (bacteria, fungi and protozoa) that inhabit an environment and can be found in different places. In almonds, the microorganisms constantly found are of the fungal genera, with Aspergillus, and Penicillium sp. being more expressive in drying, and in fermentation, yeasts are responsible for the breakdown of glucose in the pulps releasing ethanol and zygomycetes [6] .
The aim of the study was to characterize by molecular taxonomy the fungal species isolated after fermentation and drying of cocoa beans.
2. Materials and Methods
2.1. Data Collection
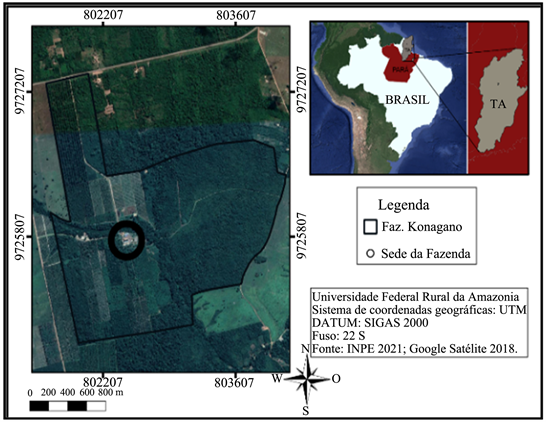
The almonds were fermented in “Fazenda Konagano”, is located in district of the state of Tomé-Açu, Pará. Afterwards, they were submitted to three types of drying, in the Laboratory of Agronomy Engineering of UFRA as shown in Figure 1.
The 10 samples of cocoa beans were collected, in the initial and final stages of each process (drying).
For drying, the samples were collected in their respective dryers at the beginning of drying in ideal conditions, then sent to the Microscopy laboratory of UFRA to identify the fungus and its incidence for each drying method used in the study [7] .
2.2. Isolation of Fungi for Morphological Identification
When evaluated the fungi, the colonies were cultivated in culture medium, Sabouraud agar (Laborclin), at a concentration of 65 g per 1000 ml of distilled water, sterilized in an autoclave for 15 min at a temperature of 118˚C [8] . Then, the sample was cultured and incubated 25˚C ± 1˚C/21days, the readings of the plates were taken at the time interval of 7, 14 and 21 days [9] .
The plates with mixed culture were frown with the fungi of interest in Potato Dextrose Agar (concentration of 18 g of dextrose, 200 g of potato and 20 g of agar, autoclaved at 121˚C for 20 min), incubated at 25˚C ± 1˚C/14days. In order to guarantee the purification of the fungus, 2 ml of distilled water was diluted in the plate where the fungus is isolated. The culture was poor in nutrient, water agar, and the fungus solution was striated by “spread plate” technique and incubated for 24 hours, thus obtaining the isolated colony.
To visualize the fungal spores and hyphae, a glass slide was prepared, the dye Methylene Blue was added, then, with the help of a needle, a fungal sample was collected and, on the plate, superimposed on a cover slip and visualized under an Optical Microscope (SP labor, Model: BLUE1000-BIL-BI) [10] .
2.3. Extraction of DNA from Fungi
DNA from bacteria was extracted using the CTAB (cetyl trimethylammonium bromide) protocol described by [11] , modified.
For DNA extraction, the fungi were initially grown on enriched and differential solid medium, Blood-Agar-(AS) with growth for 36 hours. After cultivation, one to three colonies of bacterial growth were added to microtubes containing 567 µL of 1× TE and vortexed. To fisicla break the cell wall of the fungi, expose and extract the DNA, 30 UL of 10% SDS, 3 µl of proteinase K were used and incubated for 1 to 1.5 hours at 37˚C, then 100 µl of 5 M NaCl, 80 IU of CT 80 µL of CTAB/NaCl placed in a 65˚C water bath for 10 minutes, 780 µL of chloroform/isoamyl alcohol and shaken manually for 10 minutes and centrifuged for 5 minutes at 14.000 rpm. Then the supernatant was transferred to new tubes and 360 µl of ice-cold isopropanol added. Then, the microtube was inverted until the DNA became visible and it was centrifuged at 14.000 rpm for 25 minutes. However, immediately afterwards, the supernatant must be discarded, the DNA washed with 1 ml of 70% ethanol and centrifuged for 3 minutes at 14.000 rpm and the DNA was allowed to dry for 20 minutes in a support laminar. The DNA was resuspended in 50 µL of 0.1× TE added with ribonuclease (10 mg∙mL−1) and incubated at laboratory temperature 25˚C for 12 hours.
After the extraction was completed, the samples were quantified in QubitTM. The amplicons were analyzed on Agarose Gel (2%). Then, the samples were diluted to concentrations of 10 to in µL−1 and stored at −20˚C.
2.4. A Polymerase Chain Reaction (PCR) for Fungi
For amplification of the genetic material (the DNA), DNA concentration was performed using the Biodrop µLite spectrophotometer (Thermo Fisher Scientific, USA). For identification, the 18S rDNA gene was applied with primers 27F (5'-AGAGAGTTGATCMTGGCTCAG-3’) and 1492R (5'-ACCTTGTTTTACGACTT-3’) with approximately 1450 base pairs of amplification.
PCR was performed with a total volume of 25 µl of a reaction mixture consisting of 12.5 µl of 2× Master Mix PCR (Promega GoTaq® Master Mix, Wisconsin, USA), 20 pmol of the primers and 100 ng/µl of bacterial DNA. PCR was performed in a thermocycling apparatus (Mastercycler, Eppendorf, Germany). PCR was performed with the following parameters: initial denaturation 4 minutes at 94˚C, followed by 25 cycles of denaturation 94˚C for 1 minute, annealing 55˚C for 1 minute and extension 72˚C for 1 minute and final extension 72˚C for 7 minutes. PCR amplification was analyzed by agarose gel electrophoresis (2% w/v). The run was performed in 0.5× TBE buffer at 120 V for 40 min. Visualization was performed with a transilluminator device and images were captured using UPV software (BioDoc-it TM).
PCR products were purified using the Exo + SAP enzyme mixture containing two hydrolytic enzymes: recombinant shrimp alkaline phosphatase (rSAP) and Exonuclease I (Exo I), following the manufacturer’s recommendations.
2.5. Fungal DNA Sequencing
For bacteria, DNA-templates were used the primers 27F (5'-AGAGTTTGATCMTGGCTCAG-3') and 1492R (5'-ACCTTGTTACGACTT-3’). The sequencing of the samples was carried out at the company ACTGene Analyses Moleculares Ltda. (Biotechnology Center, UFRGS, Porto Alegre, RS) using the AB 3500 Genetic Analyzer automatic sequencer equipped with 50 cm capillaries and POP7 polymer (Applied Biosystems). The DNA-templates were purified with the ExoSAP-ITTM PCR Product Cleanup reagent (Applied Biosystems) and quantified in the Nanodrop 2000 c instrument (Thermo Scientific). Afterwards, they were labeled using 2.5 pmol of specific primer and 0.5 μL of BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) reagent in a final volume of 10 μL. Labeling reactions were performed in an LGC XP Cycler thermocycler with an initial denaturation step at 96˚C for 3 min followed by 25 cycles of 96˚C for 10 sec, 55˚C for 5 sec and 60˚C for 4 min. Once labeled, samples were purified by precipitation with 75% isopropanol and washing with 60% ethanol. Precipitated products were diluted in 10 μL of Hi-DiTM formamide (Applied Biosystems), denatured at 95˚C for 5 min, cooled on ice for 5 min and electro injected in the automatic sequencer. Sequence data were collected using Data Collection 3 software (Applied Biosystems) with parameters Dye Set “Z”; Mobility File “KB_3500_POP7_BDTv3.mob”; BioLIMS Project “3500_Project1”; Run1 Module “FastSeq50_POP7_50m_cfv_100”; and Analysis Module 1 “BC-3500SR_Seq_FASTA.saz”.
The resulting Data Collection files (.ab1; electropherograms) were converted to FASTA files (.seq; text) by Sequence Analysis Software v. 6 (Applied Biosystems) under default parameters. These sequences were compared to with 18 species rDNA gene sequences for fungi available in the GenBank database, the blast algorithm (http://www.ncbi.nlm.nih.gov/Genbank/index.html). Multiple sequence alignments were performed using ClustalW (Kyoto University, Bioinformatics Center; http://www.genome.jp/tools/clustalw/ ).
2.6. Phylogenetic Analyses
Phylogenetic relationships among the fungi strains were performed by the alignment of sequences using the Clustal X 2.0 software [12] . The Unweighted Pair Group Method with Arithmetic Mean (UPGMA) phylogenetic trees were constructed using the software Mega 11 [13] .
3. Results
3.1. Morphology Analyses
The fungi most found in the drying of yards were Aspergillus spp. and Penicillium spp. Morphological characterization, Figure 2, for being suggestive fungi, for being xerative fungi, for low moisture content to avoid deprivation, must be in accordance with [6] .
3.2. Taxonomy Molecular
Molecular Taxonomic Identification of Fungi Isolated from Cocoa beans.
An identity above 80% was found in all Indians, being considered reliable for molecular identification, Table 1.
![]()
Figure 2. Fungal growth on BDA agar culture medium. R2: Macroscopy Aspergillus (suggestive); R2: Microscopy Macroscopy Aspergillus spp; R2 and R3: Yeast Macroscopy; R2 and R3: Yeast Microscopy (suggestive); R4: Penicillium Macroscopy (suggestive); R4: Penicillium microscopy (suggestive).
![]()
Table 1. Identify of Fungy present in cocoa beans after drying.
Fonte: Os Autores (2021).
The species found in this study: Geotrichum candidum and Galactomyces geotrichum are endophytic fungi. On the other hand, Ascomycota sp. and Cladosporium cladosporioides are environment fungi (Table 1).
3.3. Phylogeny DNA
The taxonomy result demonstrates the proximity relationship between all analyzed species.
The studied sequences showed similarities with sequences deposited in a database. Most of them shared the same phylogenetic clade. Galactomyces geotricum R1 was similar to Geotrichum candidum MW493646.1. Cladosporidium species was similar in its accession number, Geotrichum candidum R3 sequences was close to Galactomyces geotricum JN903644.1 and Ascomycota sp. KM458803.1 was similar to its sequence in accession number (Figure 3).
4. Discussion: Molecular Taxonomy and Phylogenetic Analyses
Geotrichum sp. belongs to the Phylum Ascomycota, Class Saccharomycetes, Order Saccharomycetales and Family Dipodascaceae [14] .
Geotrichum candidum is the anamorph (sexual reproduction) of Galactomyces geotrichum, formerly called Galactomyces geotrichum [15] [16] .
Geotrichum candidum, this yeast was found after drying, secretes hydrolytic enzymes such as pectinolytic enzymes, which help in the aeration of the fermentation, liquefying the husk and favoring the growth of bacteria. These enzymes are important in the formation of chocolate aroma, for the food industry, for the textile industry and for biotechnology in the synthesis of compounds with high added value [17] .
According to [18] , the yeast Galactomyces geotrichum used in his research as a biotechnological potential in the initial fermentation process, showed to have promising characteristics in the production of protease and toxins. Some products antimicrobials are lethal to other more sensitive yeasts, promoting competition for interference, this characteristic is important for the food and beverage industry.
In the study [19] comments, the endophytic fungus identified at the Ascomycota sp. phylum level has herbicidal characteristics in seeds and antimalarials. The study of these metabolites is extremely important for the pharmaceutical industry in drug development.
Cladosporium cladosporioides is specie of the Phylum Ascomycota. Cladosporium cladosporioides is an omnivorous fungus, considered to be saprophytic or weak opportunistic pathogen.
Other endophytic fungal species which the genus Trichoderma spp., such as T. harzianum and T. viride species that show their ability to act as a disease control agent in various cultivated plants, Fungal species may attract research attention due to their versatility of action, for be able to produce antifungal and highly competitive substances in the environment [20] .
The researches [21] found endophytic fungal species such as Penicillium indicum; Penicillium citreonigrum; and Petriella sordid, Penicillium chrysogenum, with inhibitory effects on the growth of gram-positive and gram-negative bacteria and phytopathogenic fungi in purslane plants growth.
The endophytic fungi have been associated in the control of antagonistic microorganisms phytopathogenic in seeds isolated in soil to increase quality, promoting the product preservation [22] .
5. Conclusion
However, the study contributed to the knowledge of some fungi: Galactomyces geotrichum, Geotrichum candidum, Cladosporium cladosporioides, Ascomycota sp. These biodiversities of fungi in cocoa were isolated in the region of Tomé Açú in the state of Pará. In addition, they were described in the literature participated in the fermentative process and presented antimicrobial characteristics.
Future studies should be carried out for the taxonomic characterization of more species present in the cocoa microbiota, as well as the verification of biotechnological characteristics of these species with the potential for biocontrol of phytopathogens.
Acknowledgements
We thank the UFRA Plant Pathology Laboratory in Belém/PA and Microscopy Laboratory in Campus Tomé-Açú/PA.
Author Contributions
Conceptualization: Raquel Soares Casaes Nunes; methodology: Jeferson Santana Quadros and Mateus Barros Cardoso; software: Raquel Soares Casaes Nunes; validation: Raquel Soares Casaes Nunes; formal analysis: Jeferson Santana Quadros and Mateus Barros Cardoso; investigation: Jeferson Santana Quadros and Mateus Barros Cardoso; data curation: Magnun Antônio Penariol; writing original draft preparation: Magnun Antônio Penariol and Raquel Soares Casaes Nunes; writing review and editing: Magnun Antônio Penariol and Raquel Soares Casaes Nunes; supervision: Raquel Soares Casaes Nunes; project administration: Raquel Soares Casaes Nunes. All authors have read and agreed to the published version of the manuscript.