1. Introduction
Phenolic compounds are ubiquitous among higher-order plants, primarily existing as lignin, a polymer composed of the monolignols p-coumaroyl, coniferyl, and sinapyl alcohols [1] [2] . Simpler forms of phenolic compounds are also quite prominent in a wide range of plants, and are known to have antioxidant roles and health benefits in human diets [3] [4] [5] . Compounds like methylcinnamate and methyl p-coumarate are often found as components in floral scents [6] . Grasses have both complex phenolic polymers (lignin) and monomeric phenolic compounds incorporated into their cell walls [7] . Ferulates in grass cell walls have been well investigated and their roles within plants are well established [8] [9] [10] . The p-coumarates (pCA) found in grass cell walls have also been studied but their role has not been well defined. Incorporation of pCA into cell wall matrices can occur in two forms: one as an ester linked conjugate to arabinofuranose (Araf) of glucuronoarabinoxylans (GAX) [11] and secondly as an ester linked conjugate of pCA and sinapyl alcohol [12] . Only the alcohol (sinapyl or coniferyl alcohol) becomes incorporated into the developing lignin polymer leaving the p-cou- marate portion to remain free and linked only by the original ester bond of the conjugate. A specific transferase has been identified that is responsible for the formation of the pCA-sinapyl alcohol conjugate (p-comuaroyl-CoA: sinapyl alcohol transferase, pCAT) [13] [14] . Recently a gene for p-coumaryl-CoA: sinapyl alcohol transferase has been identified from rice (Oryza sativa) and the expressed protein in an E. coli system showed a strong preference for p-coumaryl CoA and sinapyl alcohol as substrates [15] .
It has been proposed the formation of p-coumaryl-sinapyl alcohol (pCA-SA) conjugates could aid in the incorporation of sinapyl alcohol into lignin resulting in what is referred to as syringyl-rich regions of lignin [16] . In corn (Zea mays L.) sinapyl alcohol is not easily oxidized by the cell wall peroxidase: H2O2 system, but pCA is readily oxidized by the same system. The oxidized form of pCA (pCA radical) can be readily transferred to sinapyl alcohol residues creating a sinapyl alcohol radical that can readily undergo radical-mediated coupling reactions with other monolignol radicals [16] resulting in syringyl-rich lignin region. Whether this is the main function of pCA incorporation into grass cell walls remains to be determined.
Following up on work identifying a p-coumaroyltransferase (pCAT) in corn led to isolation and characterization of this enzyme and ultimately the identification of the putative gene for pCAT. An RNAi construct of the gene was produced and used to genetically modify corn. To test the hypothesis that pCA-SA could alter lignin composition, the gene was inserted and expressed in Medicago sativa L. Would this lead to an accumulation of pCA in Medicago cell walls and would this influence digestibility?
2. Materials and Methods
2.1. Plant Culture
Alfalfa was grown in the greenhouse (under high pressure sodium lamps, 14/10 day/ night regime, 25˚C - 35˚C). Plants were harvested at the late bud to early flower stage of development, frozen in liquid nitrogen, and stored in a −80˚C freezer until analyzed. Leaves and stems were stored separately.
2.2. Vector Cloning and Plant Transformation
The maize pCATcDNA expression vector was generated using Gateway® LR Clonase™ II enzyme mix (Life Technologies) as previously described [17] to recombine the cDNA-containing pDONR221-pCAT [13] and pMLS509 (aka pJCV86) [13] , a constitutive expression vector using the cassava vein mosaic virus (CsVMV) promoter to control transcription [18] [19] . The resulting pMLS509-pCAT expression plasmid was verified by restriction enzyme digestion and DNA sequencing. Both pMLS509-pCAT and empty pMLS509 were transformed into Agrobacterium tumefaciens strains LBA4404 and AGL1. The resulting strains were used for Regen SY alfalfa callus transformation according to published procedures [20] [21] with kanamycin selection at 50 mg·L−1 medium.
2.3. Transgenic Plant PCR Screening
Leaf genomic DNA (gDNA) was isolated based on the procedure of Rancour et al. [17] . Multiplexed PCR screening of plants for the neomycin phosphotransferase II plant- selection marker and a NADH-dependent glutamate synthase, a general gDNA marker, was performed using primers RH196, RH197, MS152, and MS153. Table 1 shows the primers used in this study.
![]()
Table 1. DNA primers used in this study.
2.4. Plant RNA Isolation, cDNA Synthesis, and Quantitative Real-Time PCR Analysis
DNA-free total RNA was isolated from 2 5 -50 mg of frozen-ground alfalfa leaf or stem tissue using the Spectrum™ Plant Total RNA kit (Sigma-Aldrich, St. Louis, MO) according to manufacturer’s protocol A supplemented with an on-column DNAse treatment. Total RNA yields and purity were calculated after spectrophotometer absorbance measurements at 260, 280 and 320 nm. First strand cDNA synthesis was performed using 1 µg total RNA and the Go Script™ Reverse Transcription System (Promega, Madison, WI) with a poly-T primer as before [17] .
pCAT primer design for quantitative real-time PCR was performed with the Roche Universal Probe Library Assay Design Center
(www.roche-appliedscience.com/webapp/wcs/stores/servlet/CategoryDisplay?catalogId=10001&tab=Assay+Design+Center&identifier=Universal+Probe+Library&langId=-1) (site link as of Sept. 10, 2013) online software using default parameters resulting in primers RH123 and RH124. Primer sequences MS786 and MS787 for Medicago sativa actin, a gene expression control, were from Verdonk and Sullivan [22] . Tissue- and plant line-specific first-strand cDNA was used for quantitative real-time PCR using BullseyeEvaGreenqPCR 2X master mix with ROX (MidSci, St. Louis, MO) based on previous methods [13] [17] [23] . Reactions were run using an ABI 7300 Real-Time PCR System using the following conditions: 10 minute denaturation at 95˚C, 45 cycles of 95˚C for 10 s and 58˚C for 1.5 min, followed by amplification product dissociation analysis. Real-time PCR data was analyzed using the LinRegPCR method [24] and software available at (as of July 23, 2013)
(http://www.hartfaalcentrum.nl/index.php?main=files&sub=LinRegPCR). N0 values for individual reactions run in triplicate were determined and used to calculate relative expression ratios and standard deviations for pCAT to MsActin expression for the indicated plant organ and plant line.
2.5. Statistical Analysis
One-way ANOVA with a post hoc Tukey test was performed using GraphPad Prism software (version 5.0f). An alpha value of 0.05 was used for all analysis.
2.5.1. Cell Wall Analyses
An abbreviated method of cell wall extraction [25] was used to prepare samples for analysis. Samples were ground in a freezer mill (SPEX 6850) after a 3 min precool and three cycles of 1 min run time and 2 min cool time at a rate of 12 CPS (impacts per second). Milled samples were transferred to pre-weighed Oakridge tubes cooled in liquid nitrogen and stored at −80˚C until further processing. Milled samples were extracted with 50 mM NaCl overnight at 4˚C followed by 30 min at 40˚C C the next morning. Material was pelleted by centrifugation at 22,000 x g (at average radius) for 20 min at 20˚C. Supernatants were decanted and pellets extracted 2 times more with 50 mM NaCl for 30 min at 40˚C. Pelleted material was suspended in 50 mM Tris- Acetate pH 6.5 and heated for 2 h in boiling water bath to gelatinize starch. Samples were cooled (~55˚C), treated with 40 U amyloglucosidase (Fluka Bio Chemika) and 20 U of 1,4-α-D-glucan glucanohydrolase (α-amylase; Sigma-Aldrich, St. Louis, MO), then incubated at 55˚C for 2 h with shaking. Reactions were terminated by adding ethanol (95%) to a final concentration of 80% and mixing at room temperature for 30 min. Samples were centrifuged as above, supernatants aspirated off and EtOH extracts properly disposed. Pellets were extracted an additional 3 times with 80% EtOH, followed with acetone washes, after which samples were stored overnight at 4˚C. The next day samples were brought to room temperature and incubated with shaking for 30 min. Material was centrifuged as before and supernatants removed. Pellets were extracted 1 time with chloroform: methanol (1:1 v/v) followed by 4 extractions with acetone, with all extractions for 30 min at room temperature while shaking. Cell wall residues were air dried in a fume hood for 2 d to evaporate organic solvents. After air drying, cell walls were placed in an oven (55˚C) for 48 hours to fully dry, and weighed to determine yields. Cell wall material was stored dry at room temperature until further use.
2.5.2. Acetyl Bromide Lignin
Acetyl bromide lignin determination was performed according to [26] [27] [28] . Cell wall samples were analyzed in duplicate for each preparation using ~25 mg accurately weighed into screw capped (Teflon lined cap) glass culture tubes (1.5 cm × 22.5 cm). Samples were maintained in an oven (55˚C) overnight before analyses. Dry samples were incubated 2 h at 50˚C in 2.5 ml 25% (v/v) acetyl bromide in glacial acetic acid. Samples were cooled to room temperature and any insoluble material remaining was cleared using a microfuge (1.5 mL of sample, 3 min, 12,000× g). Clarified supernatant (0.5 ml) was transferred to a glass scintillation vial containing 9.5 ml of 0.42 M NaOH, 18.4 mM hydroxylamine, and 12.4 M acetic acid. Absorbance scans from 350 - 250 nm were taken and the absorbance at 280 nm was used to calculate sample lignin content. The extinction coefficients used for calculations were 15.302 g−1 L·cm−1, for alfalfa cell wall samples and maize stock cell wall standard [28] . The extinction coefficient used for alfalfa samples was the average of values determined for alfalfa stems obtained from purified HCl-dioxane lignin preparations [28] . Duplicate cell wall preparations were analyzed and data were compiled according to transformation events.
2.5.3. Ester- and Ether-Linked Phenolics
Cell wall ester-and ether-linked phenolic moieties were determined using the sequential analysis as described [29] . Approximately 70 mg of dry cell wall material per sample was accurately weighed into reaction vessels for analysis. Ferulic acid dimers (diferulic acid, DFA) presented represent the sum of all forms detected, which include: 8-8¢-DFA (aryltetralin), 8-8¢-DFA, 8-5¢-DFA. 8-O-4¢-DFA, 8-5¢-DFA (benzofuran), 5-5¢-DFA, 8-5¢-DFA (decarboxylated), and 4-O-5¢-DFA. Ester- and ether-linked phenolics were identified and quantified as trimethylsilane derivatives (40 μL TMSI, Pierce and 10 μL pyridine) by GLC-FID (HP6890) on a ZB-5ms column (Phenomenex, Zebron 25 m × 0.25 mm, 0.25 μm film). The GC conditions were: injector 315˚C, detector 300˚C, and a temperature program of 220˚C 1 min, 4˚C min−1 to 248˚C held 1 min, followed by 30˚C min−1 to 300˚C before holding for 16 min. All GC temperature programs were run at 20 psi constant pressure and split ratio 35:1. Cell wall preparations were analyzed in duplicate and data was compiled according to plant accession, developmental stage, and tissue type based on two biological replicates. For seedling tissue and callus, error in data represents the standard deviation of analytical replicates.
2.5.4. NMR Analysis of Cell Walls
Alfalfa isolated cell walls were analyzed using the gel-state 2D NMR spectral analysis technique of Hoon and Ralph [30] . Briefly, ball milled cell walls (70 mg) were suspended in DMSO-d6: pyridine-d5 (4:1, 0.5 mL) with sonication (2 - 4 h) to achieve uniform swelling and suspension. NMR experiments were performed using a 500 MHz (DRX-500) Bruker Biospin instrument (Bruker, Billerica, MA, USA), equipped with an inverse gradient 5 mm TXI1H/13C/15N cryoprobe. The central DMSO solvent peak was used as the reference peak (δC 39.5, δH 2.49). An adiabatic HSQC experiment (hsqcet- gpsisp) [31] was performed, with the following parameters: 32 transient spectralincrementswere acquired from 10 to 0 ppm in F2 (1H) using 1000 data points, from 200 to 0 ppm in F1 (13C) using 320 data points of 776 NS (F1acquisition time 6.3 msec), with a total acquisition time of 24 h 19.2 min. Processing used matched Gaussian apodization in F2 and a squared cosine bell in F12D 13C-1H HSQC correlation spectra collected on Bruker Assignments are based on model compound data in the NMR database of Lignin and Cell Wall Model Compounds [32] .
2.5.5. Total Cell Wall Digestibility
Alfalfa stem digestibility was measured using the gas pressure method described by Weimer et al. [33] . Methods were modified to use 10 mL serum vials (calibrated as described by Weimer et al. [33] ) and using a sampleof 20 mg accurately weighed to 0.01 mg into 10 mL serum vials. In vitro gas production was measured with a hand held pressure meter (Model DPG1000B15PSIG-5, Cecomp Electronics, Libertyville, IL) at 0, 4, 8, 12, 24, 36, 48 and 96 h. All sample incubations and gas measurements were in a warm room maintained at 39˚C [33] .
Gas production data were fit using PROC NLIN in SAS v9.2 to the first-order kinetic model
Equation 1: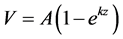 ,
,
where V = cumulative mL of gas (g of digestible organic matter)−1;
A= asymptotic mL maximal gas production (g of digestible organic matter)−1;
k= first-order rate constant; and
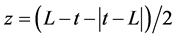 , where t = time (h) and L = discrete lag time (h).
, where t = time (h) and L = discrete lag time (h).
For all incubations, the model fit yielded a value for L that approximated zero (≤10−8), and the model equation reduced to Equation 2: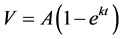 .
.
After completion of each 96 hour digestion samples were analyzed for total VFA production by HPLC [34] . Total VFA concentrations were the sum of C2 to C5 strai- ght-and branched-chain VFA. The energy content of the VFA was calculated as total alkyl groups (i.e., total methyl and methylene groups within the VFA).
3. Results and Discussion
To address the functional role of pCA in plant cell walls, we chose to stably express the maize p-coumaroyl-CoA: hydroxycinnamyl alcohol transferase in alfalfa (Medicago sativa), an agronomically important crop plant that lacks lignin-associated endogenous ester-linked pCA. In principle, this experimental system can be viewed as a gain-of- function model system. pCAT transgene expression was mediated by the cassava vein mosaic virus (CsVMV) transcription promoter [18] [35] [36] (Figure 1).
To assess the efficacy of our expression construct, the levels of pCAT gene expression were determined using real time PCR. Organ-and plant line-specific first-strand c- DNAs were generated and used as inputs. Expression of the pCAT transgene was evaluated relative to endogenous Medicago sativa actin (MsActin) expression. Using LinRegPCR software [24] , the starting values of template input (N0) into the reaction were calculated. The mean ratio of pCAT to MsActin was determined and is presented in Figure 2.
Relative expression of the pCAT transgene was detected in both leaves (Figure 2, panel A) and stems (Figure 2, panel B) in numerous pMLS509-pCAT lines. No pCAT was detected from cDNAs generated from transgenic empty vector control (EVC) plant lines. Expression of the pCAT gene in leaves extended over a range of 0.718 for line 190-3-B7 to 0 for line 190-5-B7. In stems, pCAT expression was strong but not as great as in leaves with the pCAT to MsActin expression ratios ranging from 0.327 for line 190-2-E6 down to 0.0013 for line 190-2-H1. Though not perfectly coincident, the expression level trended such that lines with high leaf pCAT expression also had high stem pCAT expression (Figure 3). The linear regression gives a line with an r2 value of 0.83 with a Pearson correlation coefficient for the dataset of 0.9115. The overall lower expression levels results in additional noise in the data. For example, lines 190-3-F1, 190-2-A1, 190-3-B7, and 190-1-C9 had high levels of pCAT expression in both leaves
![]()
Figure 1. T-DNA map for the pMLS509-pCAT expression vector. LB, left T-DNA border sequence; NOSt, nopaline synthase trans- cription terminator; NOSp, nopaline synthase transcription promoter; CsVMVp, cassava vein mosaic virus transcription promoter; attB1 and attB2, recombination sites, ZmpCAT, Zea mays p-coumaroyl-CoA: hydroxycinnamyl alcohol transferase encoding gene; T35S, 35S trans- cription terminator; RB, right T-DNA border sequence.
![]()
Figure 2. pCATgene expression in transgenic alfalfa lines Real-time PCR analysis of pCAT gene expression showing the relative gene expression of pCAT N0 to actin N0 for select alfalfa lines and tissues. The plant identifications are based on genetic background [pMLS509-pCAT vector transgenic lines (190-‘s and 188-‘s) and empty vector transgenic lines (EVC; 191-‘s)] and tissue (leaf and stem separated at harvest) used for RNA isolation. Analyses were run in triplicate with SEM error bars shown.
and stems, where as 190-2-H1, 190-5-A6, and 190-1-D5 had low levels of expression. The line 190-5-B7 exhibited a unique expression pattern where pCAT expression was detected in stems but not in leaves. The cause of this alteration was not further investigated. In summary, we were able to generate numerous transgenic alfalfa lines with broad ranging expression levels of the maize pCAT gene.
Alfalfa plant stems and leaves were analyzed separately for major cell wall components. Of particular interest was the impact upon pCA and lignin within cell walls of leaf and stem tissues. Within the stem cell wall fraction pCA was detectable at a background level of approximately 0.2 g·kg−1 CW. Variable levels of corn pCAT gene expression resulted in a wide variation in pCA incorporation into stem CW. Additional pCA incorporation above background varied from zero to nine times the background resulting in a significant increase in the ester linked pCA in stems compared to control
![]()
Figure 3. Correlation of the maize pCAT gene expression in alfalfa stems compared leaves. Linear regression gives a line with an r2 value of 0.83 with a Pearson correlation coefficient for the dataset of P = 0.9115.
plants (Figure 4). Although there were significant increases inpCA content the level was lower than what is seen in typical grass stem tissue. The pCA content of grass cell walls varies widely depending upon thespecies ranging from 5 g to 37 g·kg−1 CW [37] .
Alfalfa leaf material on the other hand already contains relatively higher levels of pCA compared to the stems. It is not clear whether this pCA is ester linked to lignin or carbohydrates. Analysis of whole cell wall material by NMR [30] does not allow us to identify pCA in the cell wall due to the limited incorporated into the wall (Figure 5). However, when comparing the total levels in alfalfa leaves to grass leaf tissue, levels are not so different from grass leaf blade tissues [38] .
This may not be too surprising because lignin levels are typically lower in leaf tissues compared to stems especially when considering the leaf tissue minus the midrib. Therefore the metabolic processes producing the monolignols will be less active than in the stems, i.e., limiting the supply of sinapyl alcohol, the preferred acceptor for the pCAT [25] [37] . If the alfalfa leaf pCA is not associated with lignin at least in the same fashion as seen with grass lignins a possibility to consider is the incorporation of pCA into the carbohydrate fraction of leaf cell walls. Mild acid hydrolysis can be used to release arabinofuranose (Araf) residues from arabinoxylans. If pCA and/or FA are attached to the Araf residue they will remain intact and one can measure the pCA-Araf- conjugate to determine the amount that is attached to at least this type of carbohydrate fraction. We have used this technique to successfully distinguish between pCA and FA bound to arabinoxylans in grasses [17] . However, this technique coupled with GC-MS analysis failed to show any pCA-Araf in alfalfa cell walls. The fact that the pCA is alkaline labile under relatively mild conditions most likely indicates it is ester linked within the wall matrix, but the wall component it is esterified to remains unclear at this time.It is possible that pCA is attached to ligninal though such attachment does not appear to
![]()
Figure 4. A modified alkaline screening method was used to look at esterified-pCA in tissue (leaf and stem) from transgenic (188-’s and 190-’s; pMLS509-pCAT) and empty vector control (191-’s) afalfa lines. Products were separated by HPLC on a C6 phenyl column and verified by standards. Alfalfa leaves compared to stems have higher endogenous levels of esterified-pCA. Expressing the maize pCAT gene in alfalfa significantly increases the esterified-pCA in its stem tissue.
![]()
Figure 5. Aromatic regions of gel-state 2D NMR spectra in DMSO-d6:pyridine-d5 (4:1). 2D 13C-1H HSQC correlation spectra are from leaf cell walls: A) pMLS509-pCAT event and B) empty-vector control and stem cell walls: C) pMLS509-pCAT event and D) empty-vector control. Aromatic rings represent unique types of aromatic units (blue-yringyl S2/6, green-guaiacyl G2 and red-p-hydroxyphenyl H2/6). Assignments are based on model compound data in the NMR database of Lignin and Cell Wall Model Compounds [32] .
be through a similar mechanism as the transferase genes and expressed enzymes seen in grasses.
Lignin concentrations in the alfalfa stems and leaves were variable both in the empty vector controls and the gene expression lines (Figure 5 and Figure 6). Overall the lignin levels were not significantly different from the empty vector controls in either stems or leaves. Some expression lines were higher and some lower but without a clear trend by expression line. Composition of the lignin appeared to shift slightly when considering the major components of sinapyl (S), coniferyl (G) and p-coumaryl (H) alcohols (Table 2) in the leaf samples.
In the stem tissue there did not appear to be any appreciable change in lignin composition (Figure 5) even though there were significant changes in the amount of pCA incorporated into the cell wall (Figure 4). The biggest impact appeared to be in the leaves, with a decrease in the ratio of S and H type lignin (Table 2) in those plants expressing the pCAT gene.
The gene is being expressed in the leaf tissues (Figure 2) and one might have predicted that there would be an increase in S type lignin in either the leaves or stems. The lack of change in stems and an apparent decrease in leaves suggest pCA is combining with sinapyl alcohol but it may not be incorporated into lignin. The decrease seen in S compared to G and H in the leaves maybe the result of conjugate formation but in
![]()
Figure 6. Acetyl bromide lignin was quantified from isolated cell walls in alfalfa tissue (leaf and stem) from transgenic (188-’s and 190-’s; pMLS509-pCAT) and empty vector control (191-’s) alfalfa lines. Error bars represent standard deviations.
leaves with a limited formation of monolignols sinapyl alcohol may become limiting. This assumes the conjugate is not ending up in lignin at least in large quantities.
We selected some of the stem tissues that showed changes in pCA content to test with an in vitro rumen fluid digestion assay. Total digestibility was measured as cumulative amount of gas produced over the 96 hours (Table 3) and by an analysis of total
![]()
Table 2. Ratios of lignin components found in alfalfa leaf and stem compared to alfalfa plants genetically modified through the expression of maize pCAT gene. Lignin components were S = sinapyl, G = coniferyl, and H = p-coumaroyl alcohols. Ratios are based upon the integration of peak areas from NMR analyses of cell walls.
![]()
Table 3. In vitro digestion analyses of genetically modified alfalfa stems. Alfalfa stem samples expressing the pCAT gene for the formation of pCA-Sinapyl alcohol conjugates were evaluated for digestibility using in vitro mixed rumen cultures. Analysis was based on single pool expon- ential model with discrete lag where A = asymptotic gas production, k = first order rate constant (reciprocal hours).
volatile fatty acid (VFA) production and total alkyl groups in the VFA (a measure of energy content) at the 96 h fermentation end point (Table 4) [34] .
Results averaged over four different replicate runs indicated there was no difference in any of these fermentation metrics between those expressing pCAT, and empty vectors as alfalfa control samples. This may indicate that pCA incorporation into the cell wall has no impact upon cell wall organization. Alternatively the changes induced within the plants by the addition of pCA conjugates were not sufficient to cause changes in cell wall structure and function, e.g., digestibility or that pCA-sinapyl alcohol conjugates do not influence total cell wall digestibility.
4. Conclusion
Modification of alfalfa by insertion of the maize pCAT gene resulted in increased amounts of p-coumarates in the cell walls. The pCAT gene expression levels were simi-
![]()
Table 4. Measurement of total volatile fatty acids (VFA) produced during the fermentation of alfalfa stem samples.
aTotal alkyl, a measure of energy content of the VFA products, is the sum of methyl and methylene groups in carboxylic acids. Mmol total alkyl was calculated from mmol of individual VFA as Acetate + (2 × Propionate) + (3 × [Butyrate + Isobutyrate]) + (4 × [Valerate + Isovalerate + 2 − Methylbutyrate]) [34] .
lar between leaves and stems resulting in similar levels of p-coumartes. However, the leaves before transformation contain relatively high levels of pCA compared to stems. Actual increases in pCA were much more pronounced in the stem cell walls with increases as high as 8 to 9 times higher than the empty vector controls. Accumulation of pCA over empty vector controls in leaves was restricted to 20% to 60% increase in those plants that increased in pCA levels. The large changes in cell wall pCA in stems did not lead to changes in lignin accumulation or a detectable shift in composition. This change in stem pCA did not result in a change in cell wall digestibility. It is possible that even though there were changes in stem associated pCA, it was not sufficient to disrupt the normal lignification process and alter digestibility.
NOTES

*Mention of a trademark or proprietary product does not constitute a guarantee or warranty of product by the USDA and does not imply its approval to the exclusion of other products that may also be suitable.