Genetic diversity of hybrid durian resulted from cross breeding between Durio kutejensis and Durio zibethinus based on random amplified polymorphic DNAs (RAPDs) ()
1. INTRODUCTION
Indonesia is a tropical country that possesses large tropical rain forests with the biggest biodiversity in the world, including genetic diversity of tropical fruits. Indonesia is one of the eight centres of plant genetic diversity in the world, especially for tropical fruits like durian [1]. That great amount of genetic diversity of Durio spp. in Indonesia’s serves an essential base for plant breeding [2].
Durian is one fruit commodities that has an important economic value. In 1985/1986, Indonesia is the second producer of durian fruit after Thailand, with the annual productivity of 200,000 tons [3]. Currently however, Indonesia imports fruits including durian from Thailand. This is because the quality of Indonesian durian is lower than Thailand durian [4].
To improve the quality of Indonesian durian, cross breeding can be done by utilizing its germplasm diversity optimally [4]. The simplest evaluation of genetic diversity is based on morphological characteristics (phenotype) which is easily observe, however it is very inadequate to differentiate genetically. This may caused by the fact that the species derived from strains from close family relationship often exhibit the same characteristics. In addition, plant breeders often find varieties with similar name but genetically may vary and are not even related, but it is difficult to distinguish phenotypically. Moreover, the phenotype is also affected by genetic factors and environmental conditions where the plant grows [5,6].
Molecular markers have become standard methods for studying variability between closely related taxa [7]. Using DNA markers the genetic relationship between samples can be estimated more accurately than the evaluation using morphology and isozymes data.
One molecular marker that has been widely used to detect variability is RAPD. RAPD method is a genetic marker in which genetic traits tend to be stable against the change and it is not affected by age. The advantage of RAPD analysis is the ability to detect differences due to the unique sequence of oligonucleotide primer corresponding to the nucleotide in the genome of the plant [8]. In addition, this technique does not require knowledge of the DNA fragments to be observed, the number of characters that can be observed is almost unlimited, fast and only requires a DNA template in low concentrations. Compared to other molecular markers, RAPD is easy to be conducted, fast, requiring a small amount of DNA (0.5 - 50 ng), does not require radioisotopes and does not require prior information of DNA sequence of the sample [9,10]. This technique has been widely used to analyze genetic diversity in crops such as citrus, sugarcane, durian sukun, grape, potato, soybean, kenaf, warty cabbage and cotton [9-17].
Cross breeding between D. kutejensis and D. zibethinus in order to generate a superior durian cultivars/ seedlings has been done in previous experiment. The advantage of D. kutejensis possesses interesting color (golden) and no smell while D. zibethinus has thick flesh. By using plant breeding technique it is expected that superior quality or production durian cultivars will arise. The hybrids were analysis using isozyme analysis. Polymorphic isozyme banding pattern of durian has been produced using esterase and peroxidase enzymes. However more molecular data of the cross breeding between the two species of durians was needed, so in this experiment RAPD analysis was done.
2. METHODS
2.1. Sample Used in This Experiment
Sample used in this study were 19 line of durian. The sample consist of 3 parent plants, one plant D. kutejensis (DRCK) and two plants D. zibethinus (DRCM1 and DRCM2), and 16 hybrids between them (UB1, UB2, UB5, UB7-8, UB10, UB13, Ub16-19, UB 21-22, UB25, and UB34-35).
2.2. Isolation of Plant DNA
Genomic DNA was extracted from 0.1 gram young leaves of durian using modified CTAB method of Doyle and Doyle [18].
2.3. RAPD Analysis and Dendrogram Construction
RAPD analysis was done using reaction mixture consist of 10 μl 2× GoTaq Green Mastermix (Promega), 3 μl of 5 μM RAPD primer, 2 μl of 20 ng/μl sample DNA and nuclease free water. The program was set at 95˚C, 15 min pre-denaturation, 45 cycles of denaturation at 94˚C for 30 seconds, anneling at 37˚C for 30 seconds and extension at 72˚C for 1 minute using 20 OPA Primer from Operon Technologies, USA with a length of 10 bases

Table 1. Sequence of RAPD primer OPA used in this experiment.
(Table 1). The RAPD results were run on agarose gel 1.0%. The amplified fragment were then converted into binnary data using Clad 97 program to analyse the similarity between lines.
3. RESULT AND DISCUSSION
Genetic Diversity of Cross Breeding Durian
From 20 primers used in this experiment, only 5 primers that capable of producing polymorphic fragments (Figure 1). The primers OPA-02, OPA-03, OPA-08, OPA-10 and OPA-13 produced 10 to 17 polymorphic fragments (Table 2).
Based on the amplification result of 19 samples DNA durian with 5 primers OPA-02, OPA-03, OPA-08, OPA- 10 and OPA-13 showed that the genomic DNA of the hybrids lines resulted from the crossbreeding of the two species of durian were divers. The DNA variation was determined according to the degree of similarity of the polymorphic fragment band resulted from PCR-RAPD amplification reaction and summarized in a form of dendogram (Figure 2).
The dendogram shows that the 19 samples which consists of three parents (DRCK = D. kutejensis, DRCM1 = D. zibethinus 1 and DRCM2 = D. zibethinus 2) and 16 hybrids lines of those parents are generally divided into 2 groups at the hypothetical taxon unit 17 at similarity value of 0.58. The first group consists of hybrids UB2,
 (a)
(a) (b)
(b) (c)
(c) (d)
(d) (e)
(e)
Figure 1. RAPD fragments of durian using. (a) OPA-02; (b) OPA-03; (c) OPA-08; (d) OPA-10; (e) OPA-13 primers. M: DNA marker of 1 Kb, (1 - 3 parents) DRCM1, DRCM2, DRCK, (4 - 19 F1hybrids) UB1, UB2, UB5, UB7, UB8, UB10, UB13, UB16, UB17, UB18, UB19, UB21, UB22, UB25, UB34, UB35.

Table 2. Number of patterns distinguished within the 19 durian result cross and varieties with different RAPD primers and total number of polymorphic fragments in the data set.
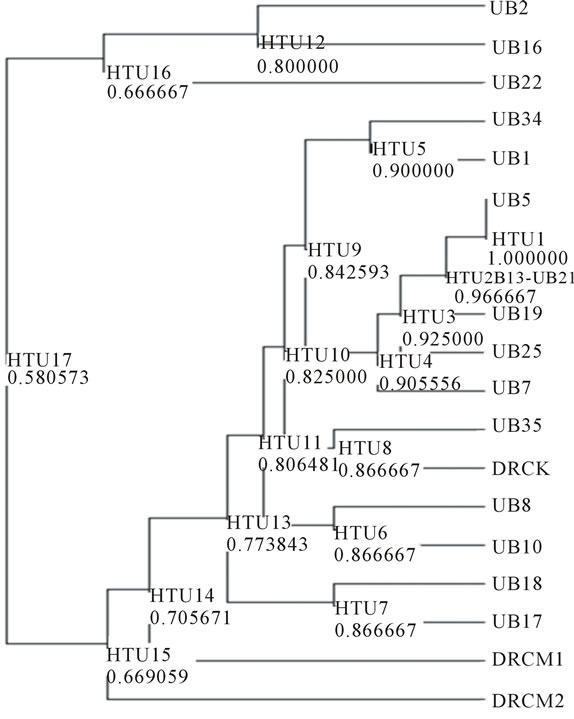
Figure 2. Dendogram generated based on polymorphic RAPD fragments.
UB16 and UB22, whereas the remaining hybrids and parents lines were grouped into the second group.
The second group consists both parents lines and all other hybrids lines. The parent lines D. zibethinus, DRCM1 and DRCM2 separated from D. kutejensis, DRCK and all other hybrid lines at similarity value of 0.71, whereas among all other hybrid lines, UB35 is the most closely related to DRCK. The other hybrid lines were related to the DRCK with similarity value ranging from 0.77 to 0.87. So it can be seen that the crossbreeding between D. kutejensis and D. zibethinus produce high DNA diversity on the hybrid lines (F1). However, it seems that D. zibethinus gave limited contribution to the diversity of the hybrid lines. Base on this finding, backcrossing the hybrid lines to D. Zibethinus parent need to be done in further experiment. Backcross procedure will help to attract more D. Zibethinus character and gain into the hybrid lines.
High genetic diversity is one of the important factors to produce the new superior varieties [19]. Increasing of genetic diversity can be done by utilizing the available of germplasm in nature and can be also achieved through crossbreeding course. So, although in this research, D. zibethinus does not seem giving a high contribution on the diversity of hybrid lines, but the cross-breeding between D. kutejensis and D. zibethinus, has been capable in increasing genetic diversity of durian lines. The increase of genetic diversity will support the genetic improvement efforts of durian [20]. In plant breeding estimation of genetic relationships are very useful for managing the diversity of germplasm and deciding appropriate lines or cultivar for crossbreeding purpose [21]. Based on the clustering showed in the dendogram (Figure 2) hybrid lines UB2, UB16 and UB22 is recommended for source line for the next crossing experiment. These lines can be crossed with all the hybrid lines or parent D. kutejensis and D. zibethinus in the other cluster. This crosses predicted to be more vigour since the source line were quite far apart which mean they possessed quite differ genetic property.
Comparing the RAPD analysis with isozyme analysis by Bansir [4], it was found that different dendogram pattern was generated. Parent DRCM which was separated from hybrid lines based on RAPD analysis, positioned in a very close relationship to UB7 and in similar cluster (group) with UB4 and UB5 based on isozyme analysis. Similarly, UB 8 which was separated from DRCK at similarity value of 0.81 based on RAPD profile, using isozyme analysis was positioned very close to DRCK at similarity value of 0.95.
These differences were presumably because PCRRAPD methods analyse at the DNA level, in which DNA is the blueprint or genetic material that carries genetic information from parent to the generation and to maintain the continuity of the structure, properties, functions and chemical activity within the cell [22].
The DNA is a genetic material or substance of the gen, whereas isozymes are enzymes that differ in amino acid sequence but catalyze the same chemical reaction [23]. This means those enzymes that catalyze the same chemical reaction could be codes by different sequence of DNA. This explained the difference pattern of the dendogram based on RAPDs and isozyme.
Genes are units of the chromosome that contain code for biosynthesizing specific protein. These proteins further determine the development of the appearance, shape, and function of tissues and organs involved. Alternative or different forms of a gene are known as alleles [24]. DNA markers can provide estimation of genetic kinship more accurately than the method of evaluation morphologically or using isozyme data [7,25]. RAPDs are able to detect genomic differences that led to the unique primary nucleotide sequence corresponds to the nucleotide in the genome of the plant.
For the next experiment, the possibility of using Restriction Fragment Length Polymorphism (RFLP) was considered. Although RFLP analysis takes much more time and needs high quality DNA, RFLP loci have multiple alleles that are readily distinguishable. Therefore more the information can be drawn from RFLPs data. RFLPs also recognized as co-dominant which capable of distinguishing between homozygote and heterozygote allele. Moreover, RFLP analysis is best due to the nonhomology of identical sized bands in RAPD analysis being scored as identical.
4. CONCLUSION
RAPDs capable of showing the genetic diversity of hybrids generated from the crossing between D. kutejensis and D. zibethinus. Three hybrids lines UB2, UB16 and UB22 show the highest distance to both parent. The rest of the hybrids lines grouped into similar cluster to the parents D. kutejensis, whereas the other parent (D. zibethinus) belong to different cluster separated from all other hybrid lines and parents.
5. ACKNOWLEDGEMENTS
This work was supported by the government of Bulungan, North Borneo and Directorate of Human Resources of the Directorate General of Higher Education (DGHE) Indonesia.

