1. Introduction
In a world literally drowned in data, Artificial Intelligence (AI) is becoming an increasingly important part of our lives. This new science at the junction of algebra, statistics, probability and computer science has diversified to meet the needs. Among the different branches of AI is Machine Learning (ML), which is used when it is difficult or impossible to define explicit instructions to give to a computer to solve a problem, but we have many illustrative examples at hand. We can oppose a classical program which uses a procedure and the data it receives (input) to produce answers (output), to a Machine Learning program, which uses the data and the answers in order to produce the procedure which makes it possible to obtain the latter from the first [1] .
AI, in general, and machine learning in particular, are progressively becoming strategic research axis for decision support solutions in several fields such as finance, marketing, security, etc. AI has also popped into agriculture and livestock, especially by contributing to improving the health and production of animals [2] [3] , but also in the field of genetic improvement and conservation [4] . This is the case of the West African taurine cattle, also known as Lobi or Baoulé. Taurine cattle are tolerant to trypanosomosis disease though smaller in size and with lower productivity compared to most zebu-type cattle [5] . Trypanosomosis is the main parasitic disease of ruminants in wetlands, causing enormous economic losses to producers. However, for the Sahel region, these wetlands are the most suitable places for livestock production because of the abundance of fodder and pasture. The effects of climate change are accelerating the phenomenon of zebu migration to these areas that were once known as taurine sanctuaries. Uncontrolled and indiscriminate crossbreeding among local cattle types is thus taking place, leading to the dilution of trypano-tolerance ability and threats to the genetic integrity of West African taurine cattle types [5] . Therefore, empirical methods of distinguishing the two species, formerly based on visual differences in morphological traits (size, presence of hump, etc.) no longer work. An efficient yet very costly method is the laboratory analysis of blood samples. Our study aims at proposing a low-cost method inspired by machine learning techniques to easily make this distinction. In the long run, it is planned to integrate the results achieved here with image processing applications to identify purebred taurines using their images.
This paper is structured as follows. In Section 2, we present the context of the problem we have to address. In Section 3, we give an overview of related work. Section 4 will provide definitions and background. In Section 5 and Section 6, we will respectively unveil some results and conduct discussions. Finally, Section 7 concludes the paper.
2. Context
There are two subspecies of cattle: zebus and taurines. The taurine cattle live in the wetlands. This fodder-rich region is unfortunately infested with tsetse flies, a vector for the spread of an endemic parasitic disease called trypanosomosis, that causes enormous losses to livestock:
· direct economic losses due to morbidity;
· stunted growth of young animals;
· weight loss;
· low milk production;
· infertility;
· abortion of cows;
· etc.
The taurines are special in that they have a natural resistance to trypanosomosis. Unfortunately, this genetic faculty is undermined by repeated cross-breeding over several generations with zebus due to the seasonal transhumance of the latter towards the wetlands and deliberate actions of breeders who seek larger animals through these crossings.
In order to preserve this type of cattle, it is necessary to find out whether a given individual is pure taurine or not. The empirical segregation methods are less and less accurate because of the massive crossings. The only formal method is a genetic analysis which is too costly in time and resources. Therefore, artificial intelligence is used for this characterization.
A conservation project working in the Sahel that focuses on the preservation of bulls has made several scientific productions on the topic, though in the field of natural and social sciences. For our study, we have in hand the data collected by this research project. Phenotypic data were measured on several thousand cattle in accordance with the 2012 FAO guidelines [6] for the phenotypic characterization of animal genetic resources (Table 1). Blood samples were also taken for laboratory analysis. These analyses allowed, among other things, to determine formally if an individual is a purebred taurine (with full trypano-tolerance capacity), pure zebu (no trypano-tolerance capacity) or a crossbred (some percentage of trypano-tolerance capacity). In the present work, we use the first dataset of 1968 individuals (taurines, zebus, crossbreds) in which six traits have been assessed: height at withers, chest girth, body length, weight, sex and age.
3. Related Work
Animal species identification is an important issue for the modernization of livestock. The scientific literature reveals different techniques that replace direct observation methods. These techniques are mainly based on body measurements, images or biological markers. One important issue is how to obtain the body features. Traditional direct measurement of animals consumes time and effort. For instance, the use of scales for live weight measurement requires a vehicle, some qualified personnel and special facilities. To overcome this difficulty, [7] and [6] used barymetric equations to estimate the weight applicable to Niger Azawak and Burkina Faso taurine cattle. This technique main drawback is that it provides low accuracy with adult animals because of the possible fattening or the
![]()
Table 1. List of quantitative traits.
physiological state of females.
Rudendko [8] derived cows’ weight using artificial neural network algorithms. This is achieved in two steps: firstly, a convolution neural network(CNN) is used to detect cows in the picture, and the stereopsis method allows the system to obtain their size measurements such as wither height, hipheight, body length and hip width via photogrammetry; secondly, these measurements are used to determine the cow live weight.
References [9] and [10] trained CNN classifiers to classify images of dogs to the appropriate class out of 120 breeds of dogs. The problem is tackled as an image classification problem using a deep convolutional neural network. In [10] , the image is divided into numerous lattices and the extracted descriptors serve as input for the CNNs that are trained to identify dog species.
Reference [11] implemented an effective breed identification system using genetic markers single nucleotide polymorphisms (SNPs) genotyped from pigmeat products. Six machine learning methods were trained to make this identification task. SVM yielded the most accurate performance.
The identification methods outlined above are based on costly techniques in computational resources as well as material resources. Our approach, which also offers good accuracy, is based on Machine Learning, using phenotypic data collected from hundreds of cows to predict their sub-species. This method can be integrated into a lite and affordable intelligent system for breed recognition in the Sahel social and economic context.
4. Methods
4.1. Conceptual Framework
The problem that we have to deal with is to decide whether a designated bovine individual is pure taurine or not. For this purpose, we dispose of its morphological measurements. To train our model, we also have at our disposal the measurements of thousands of other individuals (examples) with their label: the “pure” character. The problem is, therefore, a supervised learning mater. According to [12] , supervised learning is the machine learning task of learning a function that maps an input to an output based on input-output pairs.
For the sake of simplicity, we will restrict our attention in this phase, to determining whether the individual is pure or not, regardless of the inter-breeding rate. So, the space of labels is binary: {pure, notpure}. We are thus reduced to a binary classification problem.
4.2. Selection of Algorithms
Machine Learning relies on different algorithms to solve data problems. Choosing an appropriate classification algorithm for a particular problem task requires practice and experience [13] . At this stage of our study, we have chosen a limited number of the most commonly used algorithms, making sure that they are as representative as possible of the different types of algorithms: linear, non-linear, instance-based, bayesian, and ensemble methods.
After a model is trained, we evaluate its performance on the test set to guarantee that future measurements in similar situations are sufficiently accurate. To compare the two models, we can compare their accuracy, precision, or recall values. Reference [14] recommends that AUC (Area Under the Curves) be used in preference to overall accuracy for single number evaluation of machine learning algorithms.
4.3. Overview of a Few ML Algorithms
In the following lines, we will briefly describe the general principles of the machine learning algorithms that we will implement in this study. We will consider the following notations:
· n: the number of examples;
· m: the number of features of an example;
·  :
: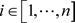 : the ith example;
: the ith example;
· 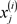 ;
; 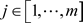 or simply
or simply 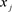 if there is no ambiguity: the jth feature of the ith example
if there is no ambiguity: the jth feature of the ith example
· 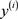 and
and 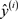 respectively the true class label and the predicted class label of the ith training example
respectively the true class label and the predicted class label of the ith training example
·  : the jth model weight.
: the jth model weight.
4.3.1. Random Forest (RF)
Decision Trees are considered to be one of the most popular approaches for representing classifiers [15] . However, they are known to have high variance and so, tend to overfit. Random forest is an ensemble method that allows to combining several trees in order to avoid overfitting. Ensemble methods apply the “wisdom of crowd” concept. This concept is based on the idea that combining many weak learners results in a performance that is far beyond the individual performance of those learners, because their errors compensate for each other. [16] proposes to build the individual trees of the forest using different variables. So at each node a number p of variables smaller than the total number is selected before applying the splitting criteria.
4.3.2. Logistic Regression (LR)
It’s a fast model to learn and effective on binary classification problems. It’s one of the most widely used algorithms for classification in the industry [17] . The basic principle is like linear regression, where the hypothesis space consists of a linear combination of the variables:
 (1)
(1)
This linear hypothesis can yield very high values as well as very low values (below zero). Logistic regression transforms this output using the sigmoid function to return a probability value: between 0 and 1. Concretely, we apply the sigmoid function:
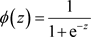 (2)
(2)
Therefore, we have:
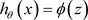 (3)
(3)
with
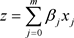 (4)
(4)
The model is trained by minimizing the cost function , using the descending gradient technique:
, using the descending gradient technique:
 (5)
(5)
4.3.3. Naïve Bayes (NB)
The model is comprised of two types of probabilities that can be calculated directly from your training data:
· The prior probability of each class.
· The conditional probability for each class given each x value.
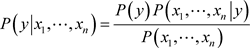 (6)
(6)
where 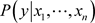 is the posterior probability,
is the posterior probability,
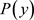 is the class prior probability,
is the class prior probability,
![]() is the likelihood,
is the likelihood,
![]() is the predictor prior probability.
is the predictor prior probability.
The predictor prior probability term is constant with regard to the class values. Therefore, we can write:
![]() (7)
(7)
The different Bayes classifiers differ mainly by the assumptions they make regarding the distribution of ![]() [18] .
[18] .
With the naive conditional independence assumption for example, this expression becomes:
![]() (8)
(8)
4.3.4. K-Nearest Neighbors (KNN)
Nearest neighbors algorithm is one of the simplest predictive models there is [13] . Predictions are made for a new data point by searching through the entire training set for the K most similar instances (the neighbors) and summarizing the output variable for those instances. We select in advance the number (K) of neighbors to consider and the notion of distance to apply. KNN is called “lazy” not because of its apparent simplicity, but because it doesn’t learn a discriminative function from the training data but memorizes the training dataset instead [17] . KNN belongs to a subcategory of non-parametric models that are described as instance-based learning.
4.3.5. Support Vector Machine Kernel (SVMk)
SVM might be one of the most powerful and widely used classifiers and can be considered an extension of the perceptron [17] . In SVM, our optimization objective is to maximize the margin that we define as the distance between the separating hyperplane (decision boundary) and the support vectors. The support vectors are the training examples that are closest to this hyperplane. The optimal hyperplane can be set as:
![]() (9)
(9)
where w is the weight vector, x the input vector and b, the bias. For all elements of the training set, w and b should verify [19] :
![]() (10)
(10)
Support vectors are those ![]() for which
for which
![]() (11)
(11)
The training objective is to find the right parameters (![]() and b) so that the
and b) so that the
hyperplane separates the data and maximizes the margin![]() . Which is equivalent to minimizing
. Which is equivalent to minimizing![]() .
.
SVM is also popular because it can be kernelized to solve nonlinear classification problems. In practice, we use a mapping function to transform the training data into a higher dimensional feature space. We now train a linear SVM to classify the data in this new feature space. The “kernel trick” allows saving expensive cost of calculations. We define a kernel function as:
![]() (12)
(12)
One popular kernel function is called Radial Basis Function (RBF) which can be written as:
![]() (13)
(13)
Here, ![]() is a free parameter to be optimized k can be interpreted as a similarity score, ranging from 0 (very dissimilar examples) to 1 (exactlysimilar examples).
is a free parameter to be optimized k can be interpreted as a similarity score, ranging from 0 (very dissimilar examples) to 1 (exactlysimilar examples).
4.4. Data Preparation
The quality of the data and the amount of useful information that it contains are key factors that determine how well a machine learning algorithm can learn [17] . Data preparation is the process of transforming raw data so that they can be run through machine learning algorithms. This involves handling categorical data and missing values, rescaling data, etc. Supervised machine learning techniques require splitting data into multiple parts for training and testing steps. However, if we are dividing a dataset, we have to keep in mind that we are withholding valuable information that the learning algorithm could benefit from. At the same time, the smaller the test set, the more inaccurate the estimation of the generalization error. Therefore, dividing a dataset into training and test sets is all about balancing this tradeoff [20] . Within the framework of this work, we used the “hold out” method. Basically, we split the dataset into two chunks: the calibration sample and the test sample. The default proportions of 70% - 30% were used.
5. Results
In the data preparation process, we cleaned the data by discarding entries that contained missing values or outliers. These inconsistent data represented 8.6% of the entire dataset. Finally, 1797 observations were validated for the study.
Data mining allowed us to visualize the shape of the feature distributions. We used pair plots to assess the correlation between the features. Graphs plotting features one against the other on the one hand and one against the label, on the other hand, showed that the interdependence is not negligible. In particular, a strong correlation was noted between weight and chest girth with a correlation coefficient of 0.948463 (Figure 1).
Correlations between the different descriptors and the “pure” trait were also analyzed. Height at withers has the highest coefficient with the label: −0.472985. This is corroborated by the feature importance graphic (Figure 3).
![]()
Figure 1. Correlation weight-girth width.
We trained the five algorithms presented earlier on the calibration set. The resulting predictive models were tested on the test sample to measure the generalization ability. Hyper parameters were adjusted to yield the best performances. The results obtained are shown in Table 2 and Figure 2.
ROC curves (Receiver Operating Characteristic) were also drawn (see Figure 4). As per [14] recommendations, AUC (Area Under the Curve)values have been calculated in order to compare the performances of the different methods.
From the AUC perspective, it appears that nonlinear Kernel SVM is the most efficient algorithm (AUC = 0.9202), followed by Random Forest (AUC = 0.9161). K-Nearest Neighbors (K = 10) and Logistic Regression have very similar performances. Their ROC curves overlap at some cut-off points and their AUC are very closed: 0.8963324783059543 and 0.8919186268624134 respectively. Naive Bayes yields the worst result. The Random Forest model remains the most accurate as far as accuracy (86%), precision (87%) and recall (88%) are concerned.
6. Discussion
Naive Bayes yields the lowest performance: 64% of accuracy. This is due to the algorithm’s strong independence assumption. It is clearly difficult to completely decouple age from chest girth or weight for example. Reference [1] demonstrated that the Naive Bayes model reaches its best performance in two opposite cases: completely independent features and functionally dependent features. In the present case, the level of feature dependence is in between.
Furthermore, the analysis of the coefficients of the regression model confirms that size (height at withers) is the most significant discriminant variable among the two species. The negative sign of the coefficients indicates inverse proportionality. This reinforces the general view that taurines are smaller than zebus. Feature importance plot (Figure 3) supports this since height at withers and body length score the most.
The strong correlation between weight and girth width can be explained by the measurement technique used in the field. Indeed, technicians used a weigh band, a tool that deduces the weight from the chest width, that is measured directly on the subject [6] .
The performance ranking showed that nonlinear models provide better results. Random Forest gives an accuracy of 86%, kernel SVM and KNN performed 85% and 83% respectively. These algorithms often lead to models with high variance. There is therefore a non-negligible risk of overfitting.
7. Conclusions
Trypanosomosis, which is prevalent in humid areas of West Africa, leads to a drop in livestock production and higher operating costs. The taurines, unlike the zebu species, have an innate ability to resist this disease. Unfortunately, uncontrolled crossbreeding between those two species of cattle leads to the dilution of this resistance capacity and threatens the genetic heritage of the taurines. Innovative means such as machine learning applications are needed to contribute to the preservation of the taurine species and its precious trypanotolerance capacity. To achieve this, it is crucial to distinguish purebred taurines from others. In this study, we applied five machine learning algorithms to train supervised models in order to make this identification. Random Forest performed the best with up to 86% accuracy, 88% recall and 0.9161 of AUC. The study confirmed that height at withers is the most discriminating descriptor among the six descriptors analyzed.
To obtain better results, it is important to continue the study by including the other morphological variables (Table 1). As this preliminary study confirmed, nonlinear methods seem to be more efficient. This trend could be further explored by the implementation of some cutting-edge models like Artificial Neural Network, XGBoost, etc. Moreover, the generalization capacity of the models trained here can be improved by associating other sampling methods such as bootstrapping.
Acknowledgements
We wish to express our sincere gratitude to all the partners who provided us with data for this study, in particular the Local Cattle Breed of Burkina Faso (LoCaBreed) team. We would like to thank Austrian Partnership Programme in Higher Education and Research for Development (APPEAR project 120). We are also grateful to Ministère de l’Enseignement Supérieurde la Recherche Scientifique et de l’Innovation (MESRSI) of Burkina Faso.