1. Introduction
The Salamandridae family, comprising 16 genera and 66 recognized species, represents one of the most diverse groups of extant salamanders. Salamandrids have the largest geographic distribution of any salamander family, extending across the Holarctic continents of Asia, Europe and North America, with a small and recent expansion into North Africa [1] .
S. infraimmaculata is present in southeastern and eastern parts of Anatolia, Turkey, northwestern Iran, northern Iraq, Lebanon and northern Israel, and has been studied during the last 40 years at the southern border of its distribution (reviewed by [2] [3] ). Based on the sequence analysis of the mitochondrial D-loop region and geological dates, Steinfartz [4] suggested that five major monophyletic groups exist in Europe (S. salamandra, S. infraimmaculata, S. corsica, S. atra and S. lanzai), one S. algira is located in Africa [5] , and the salamanders in Israel belong to S. infraimmaculata.
The populations of S. infraimmaculata found in Israel are located in breeding sites in three different and isolated sub-regions within relatively short geographical distances: 1) Mount Carmel, the southern area (SA); 2) the Galilee (Upper Galilee, Lower Galilee), the central area (CA); and 3) the foothills of Mount Hermon, the northern area (NA) [6] . The breeding sites include ephemeral pools and permanent water bodies such as streams, springs, rain pools and rock pool holes (pits), where water is available during different time periods and under various conditions.
Isolation (both initially geographic and subsequently genetic) of a population is one of the necessary conditions for evolution [7] . Geographic isolation is perhaps the most common reason for genetic divergence and depends heavily on the mobility of the organism. For example, populations of salamanders separated by large distances or by barriers (natural/humane) may never have the chance to exchange genetic material. This may lead to a faster divergence of the organisms. If geographically distant populations are genetically close, then 1) more time is needed after the separation of the populations, 2) the similarity of the distant sites leads to similar selective pressures, 3) a long-term absence of barriers to gene flow exists, and 4) there are high levels of migrants per generation, which is consistent with rapid expansion (depending on the ability of movement) [8] .
The higher diversity suggests 1) random mutation, 2) larger geographic separation, 3) separation over time between populations, 4) relatively stable isolating mechanisms acting on the population, and 5) the habitat’s climate gradient, and different biotic and abiotic conditions between habitats [8] .
The genetic consequences of habitat fragmentation are an important component of population extinction risk assessment for threatened and endangered species. Isolated small populations in fragmented habitats may undergo genetic drift and inbreeding, resulting in a decrease in allelic heterozygosity and a loss of rare alleles, and this loss of genetic diversity will increase the extinction risk of small populations [9] but presents an alternative view of polymorphism in the subterranean mammal, Spalax ehrenbergi [10] . Consequently, the estimation of genetic diversity is critical to management and conservation initiatives of concerned and threatened species and populations [11] .
Intensive studies have been made seeking genetic diversity among various isolated populations of S. infraimmaculata in northern Israel at the southernmost edge of its distribution [2] where anthropogenic pressure (e.g., tourism, housing developments, pollution, and widening of paved roads) is leading to its rapid demographic and geographical decline.
No variation has been found among populations of these areas regarding dorsal color patterns and serum proteins [12] , or in the 10 enzyme systems with 14 loci [13] , Salamanders from SA and CA habitats are significantly larger than those from NA possibly by resisting drought [14] [15] , and the molecular DNA variation using Randomly Amplified Polymorphic DNA polymerase chain reaction (RAPD PCR) found genetic variations between populations in these sub-regions but not between populations in the SA and CA habitats [16] . Goldberg [17] , who studied the same method (RAPD PCR) but examined more habitats, suggested that the genetics of populations from stable breeding sites (relatively constant water temperatures) differed from that of populations from other less predictable breeding sites. Using mtDNA sequence analysis, D-loop regions and the cyt b fragment, two subpopulations were identified among those populations. One consists of larvae from unpredictable breeding sites, of which the most common are winter pools, and the other includes populations from perennial water sources, mostly streams and springs [18] .
Our study examines genetic divergence and variation in S. infraimmaculata populations at the southernmost edge of its distribution within relatively short geographical distances using Amplified Fragment Length Polymorphism (AFLP) data. By documenting levels of molecular diversity between and among populations, we sought to answer the question if relatively short geographical distances or the extreme habitat conditions are correlated with patterns of genetic differentiation between populations.
2. Materials and Methods
2.1. Sites Studied
Ten perennial water sources and unpredictable breeding sites of S. infraimmaculata populations, covering most of its distribution range in Israel over a distance of 84 km, were examined (Table 1).
The water bodies include perennial water sources, mostly springs and streams [Balad spring (Sp1), Humema spring (Sp2), Navoraya spring (Sp3) and Tel-Dan stream (St1)], unpredictable breeding sites including winter pools [Manof pool (Po1), Dovev pool (Po3), Sasa pool (Po6), Fara pool (Po7)] and rock pool holes [Maalot pit (Pi1), Nimrod pit (Pi2)].
2.2. Larvae Capturing
Once every two weeks, 5 - 7 tadpoles of each species from a depth of approximately 10 - 120 cm were captured by a dip-net at any time in three different points in the pool (total measurement of about 15 individuals from each species) [19] . Larvae were identified, their full length was measured by a caliper (±0.5 mm), and each tadpole was released immediately at the point of capture without marking.
2.3. Collecting Samples
All samples were obtained from dip-net captured larvae (collected under Permits 2004/20306, 2005/24708, 2006/27319, 2007/30820 issued by the Israel Nature and National Parks Protection Authority). An approximately 1 cm piece of tail tissue was removed for genetic analysis and stored immediately in 95% ethanol until DNA extraction processing. Specimens were released alive in their capture location.
2.4. DNA Extraction
Total genomic DNA was extracted from clipped or whole tail of larvae with the QIAamp DNA Mini Kit (Qiagen, Germany) that consists of proteinase K lysis and specific DNA binding to the QIAamp silica gel membrane through which contaminants pass. The quality of DNA was determined by OD readings (260 nm) and by running the examples through agarose gel electrophoresis (0.8%).
2.5. AFLP Amplification
The AFLP protocol was carried out following the procedure described by Vos [20] with some modifications. High-quality genomic DNA (0.5 µg/µl) was digested with a pair of restriction enzymes (EcoRI/MseI) at 37˚C for 3 hours and then ligated to double-stranded EcoRI and MseI adaptors using T4 DNA ligase. The

Table 1. Characteristics of habitats in which salamander larvae were found.
m.a.s.l.—meters above sea level; Sp—Springs; St—Streams; Pi—Rock pool holes; Po—Pools.
double-stranded adaptor pairs were constructed from the following complementary single-stranded oligonucleoides: EcoRI adaptor: 5’-CTC GTA GAC TGC GTA CC-3’ and 5’-AAT TGG TAC GCA TAC-3’. Ligation was performed for 10 hours at 37˚C. Following ligation, each solution was diluted 10 times by sterile double-distilled water. The diluted restriction-ligation fragments were pre-amplified with nonselective primers. Pre-amplification was carried out with primers containing one selective nucleotide cytosine and adenine for MseI and EcoRI primers, respectively. PCR was performed for 24 cycles that consisted of 30 sec at 94˚C, 30 sec at 56˚C, and 1 min at 72˚C. The resulting PCR products were diluted 10 times with sterile double-distilled water and used as templates for the selective amplification step. This was carried out with a couple of selective primers (EcoRI/MseI) containing three selective nucleotides on the side that EcoRI was labeled with fluorescent dyelabeled primers (PE, Biosystems, Foster City, Califorlia).
Selective primer combinations were used for AFLP selective amplification, which was performed in two steps in a total volume of 20 μl containing 5 μl of diluted pre-selective PCR product, 1 μl of EcoANN primer at 2 pmole/μl, 2.5 μl of MseCNN primer at 10 pmole/μl and 10 μl of Reddy mix. PCR amplification cycles started at an annealing temperature of 65˚C, after which the annealing temperature was lowered by 0.7˚C per cycle for 12 cycles (a touchdown phase of 13 cycles), and then followed 24 cycles at an annealing temperature of 56˚C. An aliquot of 1 μl of selective amplification products was mixed with 8.3 μl of formamide and 0.3 μl of size standard (Liz3130).
A total of four selective primer combinations were used for AFLP amplification as follows (Table 2). The mixture was then loaded and separated on a 3130XL Genetic Analyzer Sequencer (Applied Biosystems, USA).
2.6. AFLP Data Analysis
An electrophoregram generated by the sequencer was analyzed using the GeneMapper version 3.7 software package (Applied Biosystem, 2004). Clear and unambiguous peaks, between 50 and 500 bps, were considered as AFLP markers and scored as present (1) or absent (0) in order to generate a binary data matrix. Gaps were analyzed as missing data.
DNA of several samples were amplified and run in duplicate to validate repeatability. The total number and percentage of polymorphic markers were determined for each primer pair used. The binary matrix from each primer combination was combined into one dataset and analyzed as phenotypes.
We used GenAlEx 6.4 to determine population genetic measures for each population including: N—the total number of alleles across all populations; Na—the mean number of different alleles; Ne—the number of effective alleles = 1/(p2 + q2); I/H’—Shannon Index, a measure of gene diversity applied for obtaining a partition of genetic variability within and between populations; He—calculated based on the frequencies of alleles expected, calculated under Hardy-Weinberg genotypic proportions (=2 * p * q); UHe—unbiased expected heterozygosity = (2N/(2N-1)) * He, where for diploid binary data and assuming Hardy-Weinberg Equilibrium, q = (1 – Band Freq.)0.5 and p = 1 – q; %P—percentage of polymorphic loci.
Other aspects of the genetic relationships between populations were made based on the information in the following subsections.
2.7. Genetic Structure
Bayesian Analysis of Population Structure (BAPS) Version 4.14 software [21] was used to conduct a hierarchical clustering analysis of the AFLP data. These analyses included clustering individuals independently of the location at which they were sampled, and haplotypes and clusters frequencies of the different populations across

Table 2 . Primer combinations and number of polymorphic loci per combination used in the analysis of AFLP [20].
Israel were plotted.
2.8. Analysis of Molecular Variance (AMOVA)
AFLP polimorphic loci analyzed from the DNA samples were loaded into Excel charts. Analysis of Molecular Variance (AMOVA) [22] was made using GenAlEx 6.4 software [23] . AMOVA is a method used to estimate population differentiation directly from molecular data. It tests hypotheses with respect to this differentiation. Types of molecular data, such as data from molecular markers (for example, RFLP or AFLP), direct sequence data or phylogenetic trees based on such molecular data, may be analyzed using this method. We used AMOVA tests in GenALEx 6.4 to examine the distribution of genetic variation at three hierarchical levels: within groups, defined either as subspecies or as the genetic clusters identified in BAPS; among populations; and among regions.
2.9. Mantel Test
In order to test the relationships between populations by distance and genetic diversity, pairwise genetic distances [24] -[26] among populations were obtained to produce matrices of genetic distances. Mantel test [27] using GenAlEx 6.4 with 9999 random permutations [23] was then used to test whether genetic distances between pairs of populations were significantly correlated with corresponding geographical distances.
2.10. Tree Construction
The level of genetic distance was estimated by Nei and Li’s distance matrix [28] using Tools for Population Genetic Analyses (TFPGA) software. The evolutionary history was inferred using the neighbor-joining method [29] . Phylogenetic analyses were conducted using MEGA5 software [30] .
3. Results and Discussion
A total of 479 polymorphic amplicons (bands) were generated from four combinations of AFLP selective primers in 93 individuals. The number of scoreable polymorphic loci per marker varied from 103 to 133 (Table 2). Individual clustering analysis by BAPS [21] divided individuals into genetic clusters. The fractional membership of each individual to genetic clusters is displayed in Figure 1(a). A dendrogram based on the genetic distance coefficient matrix of all genotypes analyzed showed genetic relatedness of all S. infraimmaculata sampled (Figure 1(b)). Cluster 5, which represented only the St1 population, is located on a separate branch of the dendrogram and differs from all the other clusters. Cluster 1 included 10 individuals (9.3%) from four populations and was represented in the southern area (SA) populations, very slightly in the central area (CA) populations and was absent in the northern area (NA) populations. It was found in springs, rock pool holes and pools. Cluster 2 included 12 individuals (12.9%) from seven populations and was represented in all the sub-regions and all types of breeding sites. Cluster 3, the most common of almost all populations, included 44 individuals from seven populations (47.3%) and was found in all different breeding types of salamanders larvae, but was absent in the two southern populations. Cluster 4 included 26 individuals (28.0%) from six populations from all of the areas studied in all types of breeding sites except for rock pool holes (Figure 1(a)).
The parameters by which differences among S. infraimmaculata populations are determined include morphology [12] [31] -[34] , plasma proteins [35] [36] and isozyme studies [6] [37] -[39] . Patterns of plasma proteins and allozymes are limited to protein variation, so it is difficult to use them to determine molecular variations within or between populations of the same species, as was done in the current study.
Analysis by molecular markers in the Salamandridae family has been studied intensively [1] [40] [41] , and a mitochondrial sequence analysis of Salamandra taxa suggests that populations from the Mediterranean coast of Turkey are very different in comparison to all three subspecies. Populations east of that coast are more similar to populations in central Turkey. Populations in Israel also stand out because of the DNA difference. The S. i. semenovi populations in Iran are similar to those in east Turkey. Finally, the populations of S. salamandra in west Turkey are more similar to S. infraimmaculata than S. salamandra [4] . Genetic differences were found among salamander populations in Israel using RAPDs [16] . These results are not in agreement with those of previous studies in which no variation was found in coloration and allozymes [6] .
The hierarchal AMOVA was performed once with the individuals grouped into 10 populations (Table 3(a))
 (a)
(a) (b)
(b)
Figure 1. (a) Assignment of individuals to genetic clusters (BAPS, (Corander, J., et al., 2006)) displayed by color. Each vertical line represents the fractional membership of an individual to each genetic cluster. Horizontal axis represents sampled populations. (b) Dendrogram from hierarchical cluster analysis shows grouping of S. infraimmaculata.

Table 3. Analysis of molecular variance (AMOVA) [22] based on 479 polymorphic loci of AFLP analysis. Groups defined as: (a) S. infraimmaculata from 10 populations; (b) S. infraimmaculata from five genetic populations groups clustered by BAPS [21].
d.f.—Degrees of freedom; SS—sum of squared observations; MS—mean of squared observations; Est. var.—estimated variance; % Var.—percenttage of total variance; P-value estimates are based on 999 permutations; PhiPT—proportion of the total genetic variance among individuals within populations.
and a second time with the individuals grouped into population groups assigned to five clusters (population 1— individuals showed membership to cluster 1, population 2—individuals showed membership to cluster 2, population 3—individuals showed membership to cluster 3, population 4—individuals showed membership to cluster 4, population 5—the one individual showed membership to cluster 5) (Table 3(b)). The results of AMOVA tests confirmed that the five genetic clusters identified by BAPS explained the distribution of genetic variation better than the 10 population designations: a small percentage of the variation could be attributed to differences between populations within clusters, while differences between populations within “subspecies” accounted for a much greater percentage of the variation.
Among the 10 populations, the mean percentages of polymorphic loci, He and I values were 48.7%, 0.147% and 0.227%, respectively. Population St1, a permanent breeding site where water is available all year round, exhibited the highest level of variability (%P, He and I values of 65.0%, 0.186% and 0.289%, respectively), whereas population Po6, the ephemeral breeding site, exhibited the lowest level of variability (%P, He and I values of 26.5%, 0.100% and 0.148%, respectively) (Table 4).
Intermediate values of polymorphic loci (39.5%, 38.8%) were found at populations Sp1 and Pi2, respectively, at the southern limit of S. infraimmaculata distribution and at the northern habitat in Israel, respectively. Population Sp2, which had samples only of cluster 3, exhibited 52.4% of polymorphic loci.
However, these sites are in the southernmost area of salamander distribution where conditions are only part of the time optimal to amphibians [42] . Thus, it seems conceivable that salamanders in this area may have to cope, at times, with unsuitable conditions otherwise not encountered by other populations inhabiting more favorable

Table 4 . Descriptive statistics analysis of polymorphic AFLP loci (GenAlEx 6.4, [23] ) of S. infraimmaculata from various breeding sites.
N—number of individuals sampled per population for AFLP analysis; N, Na, Ne, I, He, UHe, %P—see Chapter 2.7.3 for AFLP data analysis.
environments in the Galilee mountains [43] [44] , and this may explain their genetic variation.
The 10 habitats studied are geographically close to each other, with minor differences in rainfall, photoperiod, or other meteorological conditions. Since the rainy season is followed by eight months of hot-dry weather, the conditions of the terrestrial habitats are of great importance. However, the semi-arid conditions at the unpredictable breeding sites, where the water source dries up during the summer, differ greatly from the moist conditions in the perennial sites, where water is available all year round. We assume that the differences in cluster frequencies specifically emphasize the differences in terrestrial habitat conditions between the three sub-regions examined (NA, CA, SA). This difference could also account for the biological, morphological and physiological conditions, and possibly be responsible for the genetic variations we found from the RAPD analysis that clustered the three springs studied into one subpopulation [45] .
Nei’s genetic distance between any two populations ranged from 0.0495 to 0.1665 variable sites (Table 5), and the Mantel test [27] analysis found a low positive correlation between Nei’s genetic distance [46] and aerial distances [47] (r = 0.14, P-Value > 0.01). Hence, the geographical distance in this relatively small area has a small effect on genetic diversity. S. infraimmaculata populations considered in this work are not very distant geographically from each other, but, in some cases, important ecological conditions exist such as natural and anthropogenic barriers separating them [18] .
Gene flow between clusters (BAPS, [21] ) shows that clusters 3 and 4 are sources of migrants and also receive gene flow, while clusters 1 and 2 may be a source of migrants but may not receive much gene flow (Figure 2). Although most individuals in Sp1 and Po1 belong to clusters 1 and 2, no gene flow exists between clusters 1 and 2.
A phylogenetic analysis, which clustered Nei’s genetic distance of the AFLP data, shows that there are two subpopulations (Figure 3 ). One subpopulation includes three populations: two of the most southern populations, Balad spring (Sp1) and Manof pool (Po1), and the third group, Sasa pool (Sp6), which is a CA population. A second subpopulation consisted of populations from CA and NA [Humema spring (Sp2), Fara pool (Po7), Dovev pool (Po3), Navoraya spring (Sp3), Nimrod pit (Pi2), Tel-Dan stream (St1)], except Maalot pit (Pi1), which is a CA population. The Tel-Dan population was found to be located on a separate branch within its subpopula

Table 5 . Nei’s genetic distance [46] (below diagonal) and aerial distance, km [47] (above diagonal) between 10 populations.
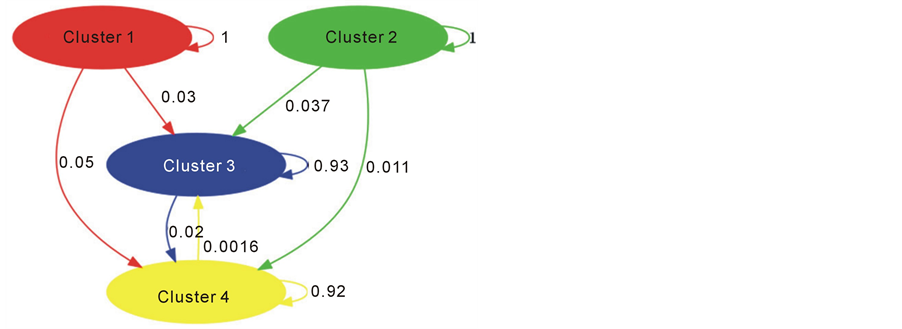
Figure 2. Gene flow between clusters (BAPS, (Corander, J., et al., 2006)). Clusters where gene flow is indicated by weighted arrows, such that the weights equal relative average amounts of ancestry in the source cluster among the individuals assigned to the target cluster.

Figure 3. Phylogenetic tree of S. infraimmaculata based on genomic DNA fingerprinting with 479 polymorphic amplicons (bands) generated from four primer combinations of AFLP. The dendrograms were drawn by the NJ algothm of the MEGA5 program using Tamura-Nei (Tamura, K., et al., 2011).
tion.
These findings are further strengthened by cluster frequencies of S. infraimmaculata that changed from north to south in a relatively small area, which was examined in the current study (Figure 4).

Figure 4. Cluster frequencies of S. infraimmaculata across norern Israel defined by AFLP data analysis (BAPS (Coranr, J., et al., 2006).
Geographic distances among the sites examined seem to have a low correlation with genetic distance, as demonstrated by the Mantel test. These findings support our hypothesis that ecological divergence prevails over geographic distance. In other words, ecological selection overrides geographic distance.
The St1 population (Tel-Dan stream( differed in the analysis from the other populations and had a unique cluster that did not appear in other populations (Figure 1, Figure 4). Exceptional ecological conditions in St1, which is a very stable habitat, i.e., with constant water temperature of 16˚C - 17˚C all year round [18] [48] , affected not only larvae growth and complete metamorphosis but also provided very stable conditions for terrestrial life of S. infraimmaculata. These conditions are extremely different regarding both larvae growth and terrestrial life compared to xeric habitat conditions found in most of the other habitats described.
However, it must be taken into account that: 1) this unique cluster is represented by a single individual of the nine individuals sampled; and 2) the Tel-Dan Nature Reserve has many creeks that are populated by salamander tadpoles, however, in this study we followed only one.
There is increasing recognition of the importance of the quality and quantity of terrestrial habitats for the conservation of juveniles and adults in the terrestrial stages [49] -[51] , and of landscape variables that potentially influence tiger salamanders movement, including distance to the nearest pond, land cover, slope and elevation [52] [53] .
The analysis of polymorphic AFLP loci of S. infraimmaculata from various breeding sites (Table 4) found that the Tel-Dan population has the highest %P and He relative to other populations of salamanders tested in this study. Our results do not concur with other studies, which found that species living under unpredictable ecological spectra generally have higher values of genetic variation than species living in more stable environments [54] [55] .
Nonetheless, we wish to put forward the following speculations about the factors responsible for this high genetic variation found in our study: the high polymorphism at Tel-Dan reflects the fact that the conditions of this habitat are stable and optimal; all the other populations examined face environmental factors that are less stable; and tadpoles subjected to optimal conditions can be very diverse and all have the potential to survive. On the other hand, extreme habitat conditions create ecological stresses and dictate certain features that must be possessed by surviving tadpoles. From the above, it is evident that additional research is needed in order to obtain a more complete and comprehensive data set to settle this apparent contradiction.
In addition to differences at the molecular level, the St1 S. infraimmaculata population was found to differ in many aspects from other relatively close populations. For example, the salamanders of St1 are smaller than the salamanders from xeric habitats [2] , are less adapted to the dry conditions [48] [56] , and exhibit a small brood size laid by each female [32] .
A relatively low gene diversity [57] is detected here (Table 4), considering the fact that habitat fragmentation and degradation of S. infraimmaculata populations have been common in northern Israel in the past few decades, and the samples used in this investigation reflect the extant gene flow among the populations.
Ecological habitats affecting genetic level were also described in S. infraimmaculata using mitochondrial and microsatellite analyses. Steinfartz [58] shows that larvae from two habitat types revealed specific adaptations and signs of genetic differentiation to these different ecological conditions.
Salamandra algira from North Africa, another population found in the southern distribution of Salamadra genus, presents a fragmentary distribution and was studied using cyt b, a control region and 12S [5] . Morphological variations among localities and genetic divergence were found among populations of the same subspecies, as was found in S. infraimmaculata and the current study.
Particularly high interpopulation differentiation within relatively short geographical distances reflected in islands, as was found in the three-island group in the southeast Agean Sea, implies long-term isolation in fragmented habitats [59] .
Habitat effect on genetic diversity in a small area was supported by other studies that examined additional amphibians species in the same area. A study of molecular DNA variation among Triturus vittatus vittatus (striped newt) from different breeding sites at the southern limit of the species’ distribution (where environmental conditions are most extreme) was made using the random amplification of polymorphic DNA (RAPD) method [60] and by variations in nucleotide sequences of the mitochondrial D-loop gene and cyt b [61] .
This saturation of different genetic diversity among the isolated populations did not just occur in the Urodela species but also in anuran at the border of its distribution. Munwes [62] found an increase in genetic variability from core to edge of distribution based on an mtDNA (control region) analysis of Pelobates syriacus from 43 rain pools distributed along a 300 km gradient in Israel. These findings were attributed to the much harsher climatic and abiotic conditions at the edge, which must be tolerated over generations by both tadpoles and postmetamorphic individuals in this region.
Gvozdik [63] studied the evolutionary relationships of tree frogs from the Middle East and the demographic histories of their populations using a combination of mitochondrial and nuclear genes. Based on genetic diversity, they suggested that there are two species in Israel. Southern populations from Yemen, Jordan, southern Syria and extreme northeastern Israel are hereby described as a new species, H. felixarabica sp. nov., and H. savignyi was found in another part.
4. Conclusion
The present study shows that the genetic divergence among isolated populations is not correlated to distance but is affected by the variation of habitats. Geographic distances among the sites examined seem to correlate less with genetic variation than ecological conditions in the habitats. The genetic divergence diversity among populations is affected not only by aquatic habitats, as was described by [18] [64] [65] , but also by adaptation to terrestrial life for which many of the adaption phenomena were studied in dilates, e.g., the number of larvae born per parturition [2] , body size [12] [35] , reproductive cycle [66] , and survival under dry conditions [48] [56] .
NOTES
*Corresponding author.