1. Introduction
Z-DNA was determined as a left-handed DNA by X-ray crystallography prior to other right-handed DNAs [1] . However, it is generally believed that under physiological conditions native DNA is mainly in the form of right-handed B-DNA. The finding of zero linking number topoisomer supports an alternative DNA structural hypothesis, the ambidextrous double helix, where there are a lot of left-handed DNAs coexisting with the righthanded DNA in the same native macromolecule [2] . Such a conceptual change would have many consequences previously unimaginable. It provided some enlightenment for various phenomena previously unexplainable. Correspondingly, it would have an impact on the theory of DNA topology and the double helix.
Actually the ambidextrous double helix is an amendment or remedy for the theory of the double helix, rather than an overthrow of its valuable core. Bidirectional winding of the two complementary strands makes the double helix more flexible and greatly alleviate the tough topological problems inherited from the Watson-Crick Model [3] . To avoid redundancy, the evidence of the finding of a zero linking number topoisomer would not be mentioned here. Interested readers may search the original paper [2] [3] .
By virtue of the wonderful resolution of topoisomers on agarose gel electrophoresis (AGE), under appropriate conditions similar topoisomers differing in just one linkage can be separated [4] [5] . Hence the band counting not only provides a way to determine the writhing number of each plasmid, but also helps scientists to find positively supercoiled DNA [6] . The finding of positive supercoiling paves the way for exploring the hidden stories of plasmids and the functions of various topoisomerases inside the cell [7] -[9] .
In this report, the helical repeat of left-handed DNA was measured experimentally. The puzzling question for the stability of hyperthermophiles chromosomal DNA at 121˚C can also be answered reasonably from a different point of view.
2. Materials and Methods
pBR322 DNA was prepared by published procedures [10] , except the clear lysate was purified with Qiagen plasmid preparation kit. Positively supercoiled DNA mixture was generated according to Lockshon and Morris [6] with their strain DK6 recA− harboring pBR322. Novobiocin, EthBr and chloroquine were purchased from Sigma Co. Electrophoresis, staining of the gel and recordings of the DNA on the agarose gel were done as before [2] . The second dimension AGE was done according to Lockshon and Morris [6] .
3. Results
3.1. A Thought Test
In 1962, the first circular DNA was discovered from polyoma virus by Weil and Vinograd [11] . Afterwards, lots of extra-chromosomal circular DNA was found from various sources, including bacteria, mitochondria, chloroplasts, etc.
Mathematician White [12] deduced a topological equation upon a closed, twisted ribbon and introduced three related elements: linking number Lk, twist number Tw and writhing number (equivalent to superhelical turns) Wr. The equation is expressed explicitly clear: . His paper is probably incomprehensible by most biochemists. However, its accuracy is well accepted and has been applied in the field of DNA topology to substitute the old equation proposed by Vinograd some years earlier, α = β + τ [13] .
. His paper is probably incomprehensible by most biochemists. However, its accuracy is well accepted and has been applied in the field of DNA topology to substitute the old equation proposed by Vinograd some years earlier, α = β + τ [13] .
Under physiological conditions, the helical repeat of a right-handed DNA is 10.4 base pairper turn [14] . Generally, all native plasmids are negatively supercoiled. According to classical DNA topology, the linking number of a native plasmid with N base pairs is less than that of their relaxed form, i.e., Lk = Tw + Wr < N/10.4. It means the two strands are underwound. Thus the negatively supercoiled DNA is equivalent to a partially unpaired DNA containing a single-stranded loop (Figure 1). It is reasonable to speculate that the size of this single-stranded loop is proportional to the absolute value of supercoiling in the negatively supercoiled DNA.
According to the traditional DNA topology, it is plausible that the higher the supercoiling of a plasmid, the lower its linking number. Theoretically, it is possible to make the plasmid carrying much higher negative supercoiling, referring to Figure 1, it is equivalent to enlarge the single-stranded loop and shorten the length of double stranded DNA. The process can be continued until it reaches to a point that the single stranded DNA loop is popped out and the two complementary single-stranded circular DNA no longer linked with each other. The test of such a thought experiment may not easy to perform, but its logic is reasonable.
In reality, this prediction conflicts with the fact that a zero linking number plasmid was found among relaxed DNA topoisomers [2] . Thus the thought experiment disproved an old notion that negatively supercoiled DNA is underwound.
3.2. The Measurement of the Helical Repeat of Left-Handed DNA
According to the traditional DNA topology, the two strands in a negatively supercoiled DNA are underwound; their average helical repeat should be more than 10.4 bp per turn.
A reasonable assumption is that the two strands in a positively supercoiled DNA are overwound. That means in a positively supercoiled plasmid, its two strands are wound more tightly and its helical repeat should be less than 10.4 base pairs.
The writhing number of monomer pBR322 is around −22 (calculated from super helical density σ ~ −0.51, Charbonnier et al. [15] ). From the two dimensional AGE [6] , it is apparent that the pBR322 obtained from a

Figure 1. Supercoiled DNA is equivalent to a partially underwound circular DNA duplex.
strain of E. coli treated with novobioc in is a mixture of negatively supercoiled and positively supercoiled topoisomers. The maxim value of positively supercoiled DNA is probably more than +22, i.e., Wr > +22.
The question is that how the two complementary strands can over wind so much, since all atoms constructing DNA (carbon, oxygen, nitrogen, hydrogen, phosphate) are not compressible and their bond length, bond angle are unable to be changed.
According to the ambidextrous double helix model, there is only one zero linking number topoisomer in the whole set of topoisomer. The linking number of all other topoisomers cannot be zero. In negatively supercoiled DNA, the linking number is a negative integer, i.e. Lk < 0, and in positively supercoiled DNA, the linking number should be a positive integer, i.e. Lk > 0. It means that the positively supercoiled DNA contains more left-handed DNA than right-handed DNA. And the zero linking number topoisomer is acting as a watershed to divide all the topoisomers of a plasmid into two groups.
It is unlikely that the two strands in those positively supercoiled DNA wound less than 10.4 bp per turn as expected from the knowledge of traditional DNA topology.
To find the answer, an experiment is carried out. By mimicking the work of Lockshon and Morris [6] with a little difference in the electrophoresis buffer, a similar 2-D gel picture is obtained as shown in Figure 2. In this picture, in the first dimension AGE buffer containing 1 μg chloroquine/ml, the topoisomer k, (the 10th band) of the positively supercoiled pBR322 DNA moves synchronously with the topoisomer L (the 11 th band) of the negatively supercoiled DNA, they were separated in the second dimension AGE with electrophoresis buffer containing 5 μg chloroquine/ml.
Suppose the helical repeat of left-handed DNA is X bp per turn and that of right-handed DNA is 10.4 + 2Y bp per turn in the buffer containing 1 μg chloroquine/ml. Here Y represents the increased value of the helical repeat due to the intercalated chloroquine molecules in the DNA. The topoisomers k and L are supposed to be a pair of morphological enantiomer which cannot be distinguished in the first dimensional AGE.
The reason of the topoisomer separation by AGE is elusive. It is generally assumed that the mobility of the topoisomer is closely related to the writhe of each topoisomer [16] . To keep the two topoisomers of opposite writhing equal, relative to the most relaxed form topoisomer which migrating the slowest at the same gel, an equation can be set up:

Likewise, in the 2-D gel of Lockshon and Morris, the 8 th topoisomer band of the positively supercoiled pBR322 DNA moves synchronously with the 9 th topoisomer band of the negatively supercoiled DNA in the first dimension buffer containing 0.5 μg chloroquine/ml.
Because the concentration of intercalating reagent is proportional to the supercoiling [4] , the helical repeat of the right-handed DNA in the buffer containing 0.5 μg chloroquine/ml should be 10.4 + Y per turn.
Another equation can be set up:
(10.4 + Y) × 9 = 8 × X
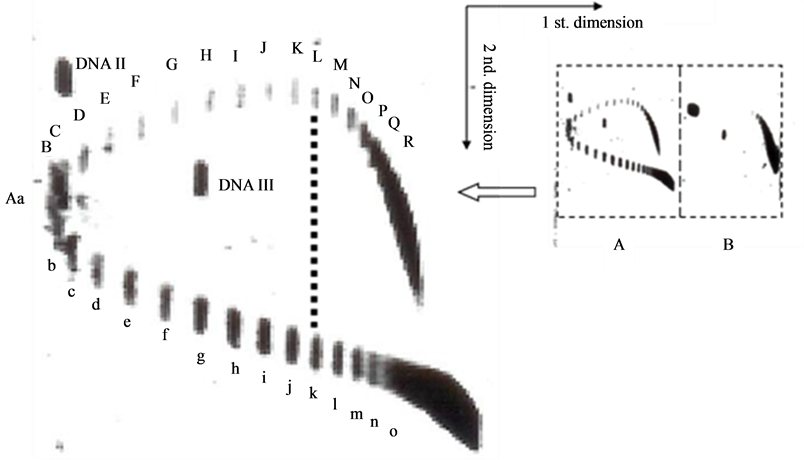
Figure 2. 2-dimensional AGE of pBR322 DNA topoisomers. The first dimensional electrophoresis is in a buffer containing 1 μg chloroquine/ml, the second dimensional electrophoresis is in the same buffer containing 5 μg chloroquine/ml. (A) novobiocin treated DNA; (B) negatively supercoiled DNA.
The answers to these two equations are: X = 11.97 bp per turn and Y = 0.24 base pairs per turn.
That means, in the buffer with 0.5 μg chloroquine/ml, the helical repeat of right-handed DNA is 10.4 + 0.24 = 10.64 bp per turn; and in the buffer with 1 μg chloroquine/ml, the helical repeat of right-handed DNA is 10.4 + 2 × 0.24 = 10.88 bp per turn. The helical repeat of left-handed DNA in pBR322 is around 12 bp per turn (calculated value 11.97). Hence the two strands in a left-handed DNA are not winding more tightly than that of a right-handed DNA.
Presently, it is not completely sure whether the chloroquine can be intercalated into the left-handed DNA. Qu et al. [17] pointed out that the ability of EthBr or actinomycin D intercalating into right-handed DNA is 300 or 1000 times more than that of Z DNA respectively. It is a reasonable assumption that the property of chloroquine is similar to that of EthBr, and very small amount of chloroquine can be inserted into the left-handed DNA.
3. Discussion
Topology is a branch of mathematics studying the spatial properties retained during deformation. The strict mathematical description makes it difficult to be comprehended by most biochemists. And the result of topological transformation is often difficult to imagine or to expect. Sometimes, even a mathematician is confused and may make some kinds of mistakes [18] .
The properties of circular DNA duplex are especially suitable for the topological investigation since the hydrogen bonds can be destroyed by various ways so that its two complementary strands are relatively un-tethered. As mentioned above, the basic topological relations were very simple as expressed in a simple equation, i.e. . However, many biochemists and molecular biochemists were unaware of the topological problems involved in the double helix. Many scientists and publishers did not notice that their published pictures were the mirror images of the right-handed double helix as revealed at a web site (The Left handed DNA Hall of Fame).
. However, many biochemists and molecular biochemists were unaware of the topological problems involved in the double helix. Many scientists and publishers did not notice that their published pictures were the mirror images of the right-handed double helix as revealed at a web site (The Left handed DNA Hall of Fame).
Karl Popper [19] claimed that except mathematics, all scientific knowledge is provisional and refutable. It is probably true that the present knowledge of the DNA topology written in most text books is not perfect. The pivotal evidence of the finding of a zero linking number topoisomer has shaken the foundation of DNA topology based on the Watson-Crick Model.
According to the traditional DNA topology, a thought experiment was designed. However, it leads to an absurd conclusion and violates the fact that the linking number of supercoiled DNA is more than that of relaxed DNA.
A peculiar and unexpected concept is that the linking number of positively supercoiled DNA is higher than that of relaxed DNA. Since the zero linking number is just between two kinds of supercoiled DNA, it leads to the deduction that the positively supercoiled DNA has more left-handed DNA. It should be pointed out that according to the ambidextrous double helix model, the left-handed DNA is extensively presented in most native DNA, unlike the Z-DNA that has a sequence requirement of alternative purine-pyramidine.
An interesting finding from the two dimension gel of pBR322 DNA topoisomers is that the helical repeat of left-handed DNA is 12 bp per turn, which is similar to that of Z-DNA. Is this a coincidence? It’s hard to tell at the moment. Anyway, science cannot prove something that does not exist or predict something that cannot be found. However, our finding may be useful when more data or facts were found by future scientist to explain the hidden secrets of DNA. It should be pointed out that the DNA structure is dynamic and polymorphous. The two kinds of DNA are interchangeable by intercalating reagents. It is probably a little different from that of B-Z DNA transition [20] . Additionally, in E. coli, probably many non-B DNA structures are playing important roles in reducing topological difficulties [21] .
Our conjecture on positively supercoiled DNA helps us to understand the stability of DNA at high temperatures.
A few years ago, the finding that certain hyperthermophilic strains can duplicate at autoclaving temperature amazed many scientists. It was reported that the doubling time of the strain 121 is 7 hr. or 24 hr. at 115˚C or 121˚C respectively [22] . The strain is still alive after staying at 130˚C for 2 hrs.
A molecular biologist would ask the question: how the DNA can resist such high temperatures without any damage, denaturation or degradation?
At first, it was supposed that the GC content of the hyperthermophilic strains may be very high, which is favorable for the stabilization of DNA. Nevertheless, sequencing data indicates the GC content is unrelated to their heat resistance.
Another speculation is that DNA is protected by some kind of histone-like proteins, such as Hta [23] or HMF [24] . The question is how much these proteins are needed to protect the entire chromosomal DNA at all times.
It was realized that reverse gyrase and positive supercoiling are always found in hyperthermophilic strains. Many scientists pointed out that both positive supercoiling and reverse gyrase are necessary for protecting DNA against high temperatures [25] [26] .
In 2004, Atomi et al. [27] found that reverse gyrase may not necessary for the survival of hyperthermophilic strain. When the unique single gene of reverse gyrase was removed from a hyperthermophilic strain, the strain is still viable at high temperatures. It clearly indicates that the reverse gyrase is not a prerequisite for the stability of DNA.
Therefore, the positive supercoiling becomes the only candidate for the heat resistance of DNA double helix. The question is how the DNA gets its positive supercoiling in the strain without the reverse gyrase. The answer may be found from the reports from Depew & Wang [28] , and Duguet [29] . They proved that the two strands of DNA are regularly turning left-handedly when the temperature is increased from 0˚C to 85˚C, and the double helix unwinding angle for each base pair is Ω = −0.01˚/˚C/bp. Charbonnier et al. [15] found this rule works up to 95˚C as well. This rule may apply at even higher temperatures. Although the unwinding value of a single base pair per Celsius degree increase is very small, the accumulated unwinding of millions of the base pairs in the chromosomal DNA of hyperthermophilic strain, together with more than 70 degree temperature difference would be big enough to transform the normal right-handed double helix into left-handed DNA.
The property of positively supercoiled DNA is governed by the law of topology. As long as the phosphodiester bonds are intact, the positively supercoiled DNA becomes a macrocosm, strong enough to resist the invasion of heat! Probably the hydrogen bonds are protected by the topological bond which is no more the only force between two atoms but is belong to the complete molecule.
The topological bond is proven effective in protecting the negatively supercoiled plasmids. At boiling temperature, plasmid DNA is rather stable, but their nicked or linear counterparts damaged or destroyed quickly. This property has been used for the preparation of plasmids in boiling method [30] .
It is reasonable to assume that both positively supercoiled and negatively supercoiled DNA is stable at high temperatures.
The positive supercoiling may be stabilized or helped by the presence of histone-like proteins or reverse gyrase in those hyperthermophiles. The evolution has its reason to set it in this way.
The concept of left-handed DNA related to positive supercoiling contradicts the traditional theory of DNA topology. The helical repeat of two kinds of DNA is different; it implied that the structure of left-handed DNA is probably not an enantiomer of that of a right-handed DNA.
Cherny and Jovin [31] reported that in low salt solution, supercoiled pPGM1 can simultaneously appear as two kinds of open configuration, with or without a 200 - 300 bp single stranded loop. DNA topology tells us that a covalently closed circular DNA with or without 200 - 300 base pairs would have a difference of 20 - 30 supercoiling which should be found under EM easily. However, the observed phenomenon defies the concept of traditional DNA topology and the classical double helix model that every 10 bp has a right-handed turn. Their results could be easily explained by the ambidextrous double helix model since simultaneous changing same number of opposite winding would not affect the supercoiling.
DNA is one of the most important macromolecules in biology and the discovery of the double helix is undoubtedly one of the greatest findings of the twentieth century. After 60 years of investigation by many scientists all over the world, lots of new findings have been accumulated. Some of them cannot be explained by the double helix, some of them even challenged the claim that all native DNAs are right-handed double helix.
Scientific theory relies on evidence. When the evidence is sufficient and enough, a rational conclusion can be reached. Just like a jigsaw puzzle, the picture will never be perfect if some parts are missing or misplaced.
When the connection is found between apparently unrelated findings or facts, it pushes scientists to explore the related fields and thus expand the frontier of our knowledge. After lots of unexpected findings and exciting breakthroughs, many secrets of life are being gradually revealed. The relation between left-handed DNA and positive supercoiling proposed here may be helpful for a better understanding of the DNA structure and its various functions.
Acknowledgements
The author is grateful to Professors H. Bremer, L. Terada, and X. Y. Liu for their support, to Dr. D. Lockshore for providing the strain DK6 recA− (harboring pBR322). Thanks to Ms. V. Poffenberger and Dr. Z. X. Shen for their careful examination of the manuscript.