Identification of a soluble phosphatidate phosphohydrolase in the developing cotyledons of Momordica charantia ()
1. INTRODUCTION
Phosphatidic acid phosphatase (PAP, 3-sn-phosphatidate phosphohydrolase, EC 3.1.3.4) hydrolyzes the phosphomonoester bond present in PtdOH yielding diacylglycerol (DAG) and Pi. Figure 1 schematically describes the reaction catalyzed by PAP. In the cytoplasmic membranes of plant seed tissue that accumulate storage triglycerides (oil), fatty acyl groups are added sequentially by specific acyltransferase enzymes to the sn-1, sn-2, and sn-3 positions of glycerol-3-phosphate (G3P) to form triacylglycerol (TAG). This pathway was discovered in the 1950s and commonly referred to as the Kennedy or G3P pathway [1]. A key step in the formation of TAG is the dephosphorylation of the sn-3 position of PtdOH, which is formed by the action of two specific acyltransferases, namely, glycerophosphate acyltransferase (GPAT) and lysophosphatidic acid acyltransferase (LPAAT). The formation of DAG is therefore the penultimate step in Kennedy pathway. The DAG is not only crucial for TAG formation, it is also important for the synthesis of phosphatidylethanolamine (PtdEtn) and phosphatidylcholine (PtdCho) [2,3]. PAP is present not only in microbes and plants but also in animals; recent studies have shown that human lipin 1 is PAP [3]. The lipin 1 deficiency in mouse prevents normal adipose tissue development which results in lypodystrophy, a disease that eventuates in the loss of body fat and insulin resistance, conversely, lipin 1 promotes obesity and insulin sensitivity [4-6]. The discovery of lipin 1 being a PAP, the enzyme is linked to the regulation of body fat metabolism in mammalian cells. Therefore, it follows that the activity of PAP will be probed by pharmaceutical researchers for the control of body fat metabolism in humans.
In the biosynthesis of TAG, PtdCho, and PtdEtn in developing seeds of oleaginous plants, the formation of DAG from PtdOH by the removal of inorganic orthophosphate (Pi) from the latter is considered a critical step [7]. Moreover, PAP has been reported as being a possible rate limiting step in TAG biosynthesis [8]. Therefore, if PAP does not function optimally, it could create a bottleneck in TAG, PtdCho, and PtdEtn biosynthesis. The kinetic and other physico-chemical data of this key biocata-

Figure 1. The schematic representation of enzymatic reaction catalyzed by phosphatidic acid phosphohydrolase (PAP). PtdOH yields DAG and Pi when phosphoester linkage is hydrolyzed by PAP.
lyst from oleaginous plants is deemed useful for further study of plant fatty acid synthesis systems and for the development of novel and alternative oil sources. This may provide an insight into further modify, enhance, and control the total fatty acyl composition of triglycerides in oil-producing plants if so desired. Of particular interest to plant biochemists are the gene sequences that codes for PAP, which may be useful for applications in genetic engineering of oleaginous crops.
In this study, we characterized Momordica charantia PAP as detected in the crude supernatant derived from developing cotyledons by measuring activity, conducting kinetics experiments, and determining pH optima, temperature optima, effects of minerals and other reagents. In mature seed of Momordica charantia, a high percentage of oil consists of α-eleostearic acid [9] which gives the oil its properties as a drying oil. Therefore, bitter gourd PAP genes could play an important role in determining high content of this industrially useful and unusual fatty acid in storage oils. This is the first documented reporting of PAP activity in Momordica charantia.
2. MATERIALS AND METHODS
2.1. Source of PAP
Momordica charantia (bitter melon) fruits at mid maturity level, which is ideal for human consumption, was purchased at an oriental grocery in New Orleans, LA, USA. The cultivar is widely grown in India and the fruits were each about 6 inches in length on average.
2.2. Extraction of Soluble PAP from Momordica charantia Developing Cotyledons
The seeds (18.0 gm) of mid-level maturity were obtained from the seed cavity of Momordica charantia fruit and washed with 0.9% ice cold saline solution. The outer coverings of the seeds were removed at room temperature using scalpel and the cotyledons removed manually. All subsequent operations were carried out at 4˚C.
The extraction buffer (5.0 ml of 50 mM acetate, pH 5.0, 150 mM sodium chloride and 10 mM MgCl2) was added to the seeds and homogenized using Tekmar Tissumizer MarkII (Cincinnati, OH) at low, medium and high speed for 30 sec each. The homogenate was cooled on ice for 1 min in between bursts. This was followed by a centrifugation using Sorvall RC2B (Miami, FL) centrifuge at 20,000 × g for 30 min at 4˚C. The pellet containing unbroken cotyledon was discarded. The resulting supernatant was dialyzed overnight against 50 mM imidazole, pH 6.0 containing 1 mM MgCl2 with three 500 ml buffer changes to remove any inorganic molecules originating from the seed tissue. The dialyzed supernatant became cloudy after dialysis, which was removed by centrifugation as above. The pellet was discarded and the supernatant was used for PAP activity measurement. The protein content of the supernatant as determined by Bicinchoninic acid (BCA) method (Pierce, Rockford, IL) was 4.27 mg/mL and the total protein was 18 mg in 4.2 mL volume. The specific activity of the preparation was 1.17 ηkat/mg of protein when the assay was performed at 53˚C and pH 6.0.
2.3. Ultracentrifugation Study of the Crude Extract
To determine whether the PAP derived from the developing cotyledons of M. charantia was a soluble protein, an ultracentrifugation experiment was conducted with the 18,000 × g supernatant fraction of the crude cotyledon preparation. The sample (4.5 ml) was subjected to ultracentrifugation (Sorvall discovery 100SE, Kendro, Newton, CT) at 105,000 × g for 1 hr. The pellet was resuspended in 4.0 mL 50 mM sodium acetate, pH 5.0, 1 mM MgCl2, buffer. Both the supernatant and resuspended pellet from ultracentrifugation study was assayed for PAP activity.
2.4. Binding of Soluble Proteins from Cotyledon Extract to Ion-Exchange Column
The supernatant from cotyledon extract of M. charantia was dialyzed against 25 mM glycine-HCl buffer, pH 3.0, overnight with three times buffer exchange, 500 ml each. The column chromatography was performed using BioLogic LP™ (Bio Rad, CA) chromatographic workstation at ambient 25˚C. The dialyzed protein was applied to a 20 ml UNOsphere™ S column (2.5 × 4.0 cm) equilibrated with the glycine-HCl buffer at the flow rate of 3.0 ml per min. The column was then washed with the equilibration buffer followed by a stepwise sodium chloride gradient in Glycine-HCl buffer ranging from 0.2 to 0.4 M at 0.05 M increment and then with 0.5 and 1.0 M sodium chloride in the glycine-HCl buffer.
2.5. Measurement of Pi Released from PtdOH by PAP Activity
To measure nmole level of Pi released by Momordica charantia PAP from the substrate, dioleoyl-phosphatidic acid (1,2-dioleoyl-sn-glycero-3-phosphate, sodium salt, Avanti polar lipids, Inc. Alabaster, Alabama) we used ammonium molybdate-acetone-acid (AMA) method [10]. Briefly, a 50 µL aliquot of supernatant containing PAP enzyme was added to 900 µL of 50 mM imidazole buffer, pH 6.0, which was kept at 53˚C in a water bath. The enzymatic reaction was started by the addition of 50 µL of substrate followed by 2 mL AMA reagent to stop the reaction at the end of enzyme assay that was typically 30 min. After 30 sec, 0.1 mL of citric acid (1.0 M) was added to each tube to fix the color. To get a clear solution, we centrifuged in an Eppendorf 5415C (Westbury, NY) at 13,000 rpm for 7 min. The absorbance was read at 355 nm after blanking the spectrophotometer with the appropriate control, which was stopped at zero time. The PAP activity was expressed as ηkat/ml (ηmoles orthophosphate released per sec).
2.6. Phosphatase Assay
A 10 µL aliquot of PAP enzyme was added to 940 µL of buffer (50 mM acetate, pH 5.0) and incubated with 1.25 mmole of ρ-nitrophenylphosphate (ρNPP) in a final volume of 1.0 mL at 55˚C for 2 min. The reaction was terminated using 0.1 mL 1.0 N NaOH and the liberated ρ-nitrophenol was measured spectrophotometrically at 400 nm [11].
2.7. Determination of pH Profile
To measure both the PAP and phosphatase activity, the choice of buffers were 25 mM glycine-HCl (pH 1.5 - 3.0), 50 mM sodium acetate (pH 3.5 - 5.5), and 25 mM imidazole (pH 6 - 9). The dialyzed seed extract was incubated in various buffers as described before to determine the pH optima of Momordica charantia PAP.
2.8. Determination of Michaelis Constant (Km) and Vmax
The Km and Vmax for Momordica charantia PAP were determined at 53˚C and pH 6.0 using the enzyme assay mentioned earlier. The concentration of dioleoyl-phosphatidic acid (DPA) ranged from 0 to 500 μM, whereas the phosphatase assay was performed at pH 5.0 and 53˚C with 0 to 1250 μM ρNPP as the substrate. WindowChem’s (Fairfield, CA) software Enzyme Kinetics version 1.1 was used to compute both the Km and the Vmax values.
3. RESULTS
3.1. Momordica charantia PAP Is a Soluble Enzyme
Both the ultracentrifugal study and chromatographic behavior of PAP in ion-exchange column have pointed out that the cucurbit PAP is a soluble enzyme. In ultracentrifugal study, the bulk of the activity (87.2%) remained in the soluble fraction. Also, the protein was bound to the S column at pH 3.0 and eluted at 0.5 M sodium chloride concentration in glycine-HCl buffer, pH 3.0 (Figure 2).
3.2. The pH Optima of Momordica charantia PAP
The pH versus activity profile of PAP for both the natural and synthetic substrates is shown in Figure 3. When DPA was used as substrate at 37˚C, the maximum activity was observed at pH 6.0 (Figure 3, panel A). However, a substantial amount of activity was seen at pH 5.0 and 7.0. Approximately 79% of the activity was found at pH 5.0 and 24% activity at pH 7.0. However, when ρNPP was used as the substrate at 53˚C, a bi-hump profile was observed (Figure 3, panel B). A minor peak was observed at pH 3.0 that constituted about 34% of activity; the major activity peak was observed at pH 5.0.
3.3. The Temperature Optima of Momordica charantia PAP
The temperature dependence of PAP activity of Momordica charantia crude supernatant is shown in Figure 4, panel A. The optimum temperature for activity was found to be 53˚C. At 50˚C, the activity dropped only about 7% and at 62˚C, about 35% drop in activity was observed. Looking at the temperature versus activity profile, one could discern that Momordica charantia PAP is a thermostable biocatalyst. The associated phosphatase activity also showed a similar temperature optima profile when ρNPP was used as the substrate (Figure 4, panel B).
3.4. The Kinetic Properties of Momordica charantia PAP
In these experiments we challenged the substrate, DPA,
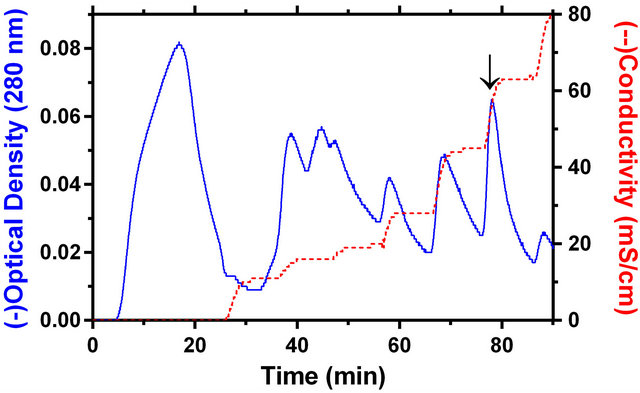
Figure 2. The cation exchange chromatography (UNOSphere™ S) of M. charantia soluble cotyledon extract. Both the optical density profile at 280 nm and conductivity of the column eluate are shown. The arrow marks the position where PAP activity was eluted by 0.5 M sodium chloride in glycine-HCl buffer.

Figure 3. The pH profile of the Momordica charantia PAP enzyme catalyzing PtdOH (Panel A), and ρNPP (Panel B).

Figure 4. The temperature versus activity profile of the Momordica charantia PAP enzyme catalyzing PtdOH (Panel A) and ρNPP (Panel B).
with the dialyzed supernatant that contained PAP at pH 6.0 and 53˚C in order to assess whether the phosphohydrolase reaction exhibits the hallmark of enzymatic reaction, viz. a sigmoidal curve for the reaction as a function of enzyme concentration and the rate linearity curve for time dependence. The amount of supernatant extract containing PAP enzyme was varied between 0 to 50 µL corresponding to 0 to 210 µg of protein and the activity was measured. In this case, a sigmoidal curve was obtained (Figure 5). In another set of experiments the incubation time of reaction was varied between 0 to 20 min. In this case, the rate linearity of the enzymatic reaction was observed (Figure 6).
3.5. Michaelis Constant (Km) and Vmax Determination for Momordica charantia PAP
When both the natural and artificial substrates were used to drive the enzymatic reaction, the enzyme gave a typical sigmoidal curve for both the substrates (Figure 7, panel A and panel B). The Km and Vmax for DPA were 142 µM and 1.89 ηkat/mg of protein, respectively. The kinetic parameters for ρNPP driven assay were computed to be 107 µM and 10.4 ηkat/mg of protein, respectively.
3.6. The Effect of Non-Ionic Detergent Triton X-100 on M. charantia PAP Activity
Triton X-100/phosphatidic acid mixed micelles was used

Figure 5. Phosphohydrolase activity of the Momordica charantia PAP as a function of enzyme concentration.

Figure 6. Phosphohydrolase activity of the Momordica charantia PAP exhibiting the time course dependence of Pi release when fixed volume of the enzyme was assayed under experimental condition.

Figure 7. The PAP enzyme activity as a function of substrate concentration. Panel A shows the hydrolysis of PtdOH and panel B shows the hydrolysis of ρNPP as a function of the concentration of substrate.
to perform kinetic studies with the membrane-bound PAP from yeast [12]. To investigate whether the non-ionic detergent would facilitate the soluble PAP from Momordica charantia in the same way, we performed PAP activity in presence of non-ionic detergent Triton X-100. The results are shown in Figure 8. The enzyme could tolerate a low concentration of Triton X-100; however at concentration ranging from 0.3% to 0.5%, the activity was decreased about 30% - 60%. The accessibility of the substrate, DPA, to the active site of the enzyme was not facilitated by the presence of the detergent.
3.7. Reaction of Momordica charantia PAP to Various Protein Sensitive Chemicals
The dialyzed crude supernatant from Momordica charantia cotyledons were treated with cysteine modifier (N-ethylmaleimide, NEM), thiol protectant (β-mercaptoethanol, β-ME), and arginine modifier (phenylglyoxal) to investigate whether these had any effect on catalytic rate of the Momordica charantia PAP. All three reagents had no effect on PAP activity (Table 1).
4. DISCUSSION
This is the first reporting of any phosphatidic acid phosphohydrolase (EC 3.1.3.4) activity in Momordica charantia, which was chosen for study because the mature seeds of the plant make TAG that contains α-eleostearic acid as the acyl chains in all three positions [13]. Maturing seeds of Momordica charantia have all the enzymes to assemble storage triglycerides (TAG); therefore, it is a perfect candidate for biochemical dissection. When the supernatant of cotyledon extracts from mid-maturity level seeds of this plant were examined for PAP activity in an in vitro assay, we detected quantifiable phosphohydrolase activity in the supernatant fraction, which was characterized and reported in this communication.
In this study, the soluble nature of Momordica charantia PAP was established by two methods, namely, the ultracentrifugation study and ion-exchange chromatog-

Figure 8. Momordica charantia PAP enzyme activity measured in presence of varying concentration of Triton X-100.
Table 1. Effect of various reactive agents on the PAP activity.

raphy. In particular, the binding of the PAP to S column and its subsequent elution from the column at 0.5 M sodium chloride in glycine-HCl buffer give credence to the notion that this protein is not a component of any membrane. There are reports of two kinds of PAP in oleaginous plants and yeasts, namely, the soluble [14] and membrane-bound PAP [15]. Based on the dependence of the biocatalyst to Mg2+ for activity, the enzyme could also be divided into 2 classes, namely Mg-dependent [6] and Mg-independent PAP [16].
Even though the biochemical evidence for PAP activity had been published over the years, the primary sequence of the biocatalyst and the nucleotide sequence of the gene coding for the protein was only discovered in 2006 from the yeast Saccharomyces cerevisiae [6]. The coded enzyme derived from PAH1 gene was reported to be associated with both the supernatant and membrane fractions of the cell; however, its association with membranes is now considered peripheral in nature [17]. The yeast PAP was reported to be specific for PtdOH and exhibited its phosphohydrolytic activity based on the active site motif DXDX(T/V) within a halo acid dehalogenase (HAD) domain Furthermore, the yeast PAH1 gene showed sequence homology to mammalian lipins encoded by Lipin1, Lipin2, and Lipin3 [4].
We have provided preliminary evidence for no participation of Arg and Cys residues in the active site of Momordica charantia PAP (Table 1). However, further research is needed to verify these findings.
The roles PAP play in lipid metabolism and cell physiology were documented through studies employing mutants defective in PAH1 encoded protein. The yeast containing the defective PAH1 gene showed elevated levels of PtdOH and decreased levels of both DAG and TAG [6, 18]. In addition, the contents of the major phospholipids PC, PE, and PI, and fatty acids were affected by the deletion mutation in PAH1 gene [6,18]. Therefore, PAP probably plays a role in the regulation of overall lipid synthesis.
The effect of non-ionic detergent Triton X-100 on Momordica charantia PAP, which was found not to be associated with internal membranes of the seed, was inhibitory. Triton X-100 forms a mixed micelle with the lipid substrate PtdOH, providing a membrane mimic for catalysis [12,15]. However, in the case of Momordica charantia PAP, the enzyme is not associated with membrane; therefore, instead of stabilizing the substrate, it probably disturbs either the substrate binding site or the catalytic site of the Momordica charantia PAP.
A thorough study on the kinetic, biochemical properties and active site mapping of the Momordica charantia PAP awaits its purification.
5. CONCLUSION
The developing seeds of Momordica charantia, the bitter gourd, were investigated for soluble phosphatidic acid phosphohydrolase (EC 3.1.3.4) activity using an in vitro assay method. We found detectable PAP activity in the supernatant fraction of the seed extract. The enzyme activity was characterized for time course, concentration dependency, pH optima, temperature optima, and susceptibility to Arg and Cys modification. The enzyme activity did not appear to be dependent on Mg2+ and the Vmax and Km were estimated to be 1.89 ηkat per mg of protein and 142 µM, respectively, when assayed at pH 6.0 and 53˚C, respectively. The enzyme was readily inactivated by non-ionic detergent Triton X-100, which indicates that Momordica charantia PAP is not a membrane bound enzyme. This PAP could serve as a model for human lypodystrophy, which is characterized by loss of body fat, fatty liver, hypertriglyceridemia, and insulin resistance.
NOTES