 Advances in Infectious Diseases, 2011, 1, 1-13 doi:10.4236/aid.2011.11001 Published Online September 2011 (http://www.SciRP.org/journal/aid) Copyright © 2011 SciRes. AID 1 HIV-1 Env gp120 C2V5 Potential N-Linked Glycosylation Site(s) (PNGs) Variations and Amino Acid Length Polymorphisms among Infected Family Members Duri Kerina1*, Felicity Zvanyadza Gumbo2, Knut Ivans Kristiansen3, Munyaradzi Paul Mapingure4, Simba Rusakaniko5, Mike Zvavahera Chirenje6, Babill Stray-Pedersen4, Fredrik Müller3 1Department of Immunology, University of Zimbabwe, Harare, Zimbabwe; 2Department of Pediatrics and Child Health, University of Zimbabwe, Harare, Zimbabwe; 3 Department of Microbiology, University of Oslo and Oslo University Hospital, Rikshospitalet, Oslo, Norway; 4Letten Foundation Research Centre, Harare, Zimbabwe; 5Department of Community Medicine, University of Zim- babwe, Harare, Zimbabwe; 6Department of Obstetrics and Gynecology, University of Zimbabwe, Harare, Zimbabwe; 7 Division of Women and Children, Oslo University Hospital, Rikshospitalet and Institute of Clinical Medicine, University of Oslo, Oslo, Norway. Email: *tkduri@yahoo.co.uk, Received August 2nd, 2011; revised September 6th, 2011; accepted September 13th, 2011. ABSTRACT Objective: To ascertain the role of HIV-1 gp120 env PNGs variations and sequence length polymorphism following transmission events as possible suppo rting forensic evidence to determine directionality of HIV tran smission. Method: An observational study of HIV-1 infected family members, where median and range values of the amino acid lengths and PNGs for the genotyped C2V5 region were calculated. Wilcoxon rank-sum test was used to determine differences in these parameters between different family members. Results: For heterosexual transmission, two mothers had longer C3 sequences relative to tha t of their spou ses; p = 0.006 and p = 0.025 whilst the opposite was observed for one mother, p = 0.028. No clear trends were observed for PNGs. In three families, index children had longer C 2V5 amino acid se- quences compared to their mothers; p= 0.013, 0.040 and 0.043. Second siblings’ V4 and V5 sequences were generally shorter relative to the maternal ones; p = 0.039 and 0.028, respectively. Generally adults had longer V3 amino acid sequences compared to the children; p = 0.018. Similar trends were also observed regarding PNGs within the entire C2V5 region, C3 and V4 sub-regions; p= 0.0025, 0.005 and 0.008, respectively. First siblings’ C2V5 and C3 sequence lengths were significantly longer relative to tho se of the second sibling s; p = 0.005 and 0.007, respectively. Conclusion: Our results are suggestive that HIV-1 env C2V5 amino acid length polymorph ism and PNGs tend to increase with age and HIV disease progression. Though sensitive and should be cautiously handled, it is tempting to propose the direc- tionality of the HIV transmission events with respect to C3 sequence length polymorphisms. Correlating HIV-1 env C2V5 amino acid length polymorphism and age of infection may be the first step towards a possible valuable piece of forensic evidence which may be useful in criminalisation of willful HIV infections. However, bigger studies are war- ranted to substantiate the authenticity of this potentially useful application. Keywords: HIV-1 Env120 C2V5, Glycosyl a tion, Amino Acid Length Polymorphism, Parent to Child Transmission 1. Introduction Human immunodeficiency virus type 1 envelope glyco- protein 120 (HIV-1 env gp120) is composed of relatively conserved constant (C) C1 to C5 and variable (V) V1 to V5 regions [1]. It is ranked one of the most heavily gly- cosylated proteins known in nature with N-linked glycans constituting over 55% of its molecular weight [2,3]. This extensive glycosylation is known to play an impor- tant role in evasion of the host immune response by masking key neutralization epitopes and presentation of the glycosylated env (glycan shield) to the immune sys- tem as “self” [4-9]. Under host immune response or anti- retroviral therapy selection pressures, it is postulated that HIV-1 evolves towards a denser glycan shield [10], an observation also supported by others [11-13]. According  HIV-1 Env gp120 C2V5 Potential N-Linked Glycosylation Site(s) (PNGs) Variations and 2 Amino Acid Length Polymorphisms among Infected Family Members to this hypothesis, shorter variant with fewer glycans are expected during earlier time points of infection whilst longer V1 - V5 forms with more glycans are expected to evolve at a later stage in response to prolonged immune pressures [14,15], making it possible to assign the direc- tionality of transmission. Regardless of the HIV-1 subtype, the number of poten- tial N-glycosylation site(s) (PNGs) within the gp120 env gene (env) is conserved at about 25 [10,16]. However, there is a tendency for heterosexually and perinatally transmitted viruses to have shorter env variable loops and fewer PNGs [14,17-21]. Changes in the number of PNGs and env gp120 V1-V5 amino acid lengths have been as- sociated with striking a balance between transmission competence and resistance to immune challenges [14,21- 23]. Whilst glycosylation within the HIV-1 env V3 loop may change the viral co-receptor usage [24], loss of PNGs within the V4 region has been correlated with de- mentia [25]. Intra-host HIV-1 PNGs diversity varies, with some PNGs being evolutionarily conserved while others may be present in some hosts but absent in others and some may even appear or disappear during the course of infection [3,10]. HIV-1 subtype B env gp120 V4 loop has been shown to appear as swarms of heterogeneous N-linked glycosylation variants due to mutations, inser- tions and deletions (indels) which consequently affect amino acid sequence lengths and PNGs distribution pat- terns [26,27]. Although glycosylation of HIV-1 remains the main obstacle to viral control and eradication, it is possible to exploit the protective glycosylation profiles of gp120 against the virus for the development of an env based vaccine candidate [26]. For this reason, understanding glycan shield glycosylation patterns is critical. There is a paucity of data in Zimbabwe regarding the number and distribution patterns of HIV-1 subtype C gp120 env PNGs and sequence length polymorphisms in adults fol- lowing heterosexual transmission, let alone in vertically infected children. More so, gp120 env characteristics of the unusually prolonged survival of HIV-1 infected but drug naïve pediatric patients are not well documented. Our study population of HIV-1 infected family members provided a unique opportunity to investigate gp120 env PNGs patterns and amino acid sequence lengths poly- morphisms following heterosexual and subsequent parent to child transmission (PTCT) events, a unique scenario that has not been reported elsewhere. 2. M at erials and Method Study Population and Procedures Described are HIV-1 transmission events of close con- tacts of whom the time and directionality of vertical transmission was known but unknown for heterosexual transmission. The unit of analysis was a family. Four families were willing to participate in our study. HIV-1 infected families labeled 205, 366, 375 and 567 consisted of biological parent(s) and children, index child (older/first sibling) and index child’s sibling (younger/second sibling), constituted the study popula- tion. The index child was defined as the first child to be recruited into our study. Two families comprised each of both parents and a respective biological index child. The other two families were composed of parent(s) and two subsequent biological children, the first and second sib- lings. For Family 567 the father figure was missing as he was working in another regional country. Each family member was HIV-1 infected and none had received anti- retroviral therapy at the time of sample collection. Consent was obtained from the respective pregnant mother of each family participating in the national PMTCT programme in Harare peri-urban mother and child clinics that were known to be HIV-1 positive at 36 weeks gestation. Spouses also consented to participate in the family HIV genetic study. Similar recruitment and procedures were followed as previously described [27]. All the infants were breast fed for at least nine months. First siblings’ blood samples were collected at 60 ± 10 months of age as there were insufficient sample volumes from their respective first available HIV-1 positive sam- ples. In cases of second siblings the first available HIV-1 positive samples were genotyped and sampling time was at about 15 ± 3 months of age. Nucleic acid extraction, PCR amplification, cloning and DNA sequencing meth- ods for the HIV-1 env gp120 C2V5 region were done as previously described and so was HIV-1 subtype determi- nation[28]. 3. Data Analysis Data were entered and analysed using Stata version 10. The number and distribution of PNGS including C2V5 sequence length polymorphisms were determined for both the parent(s) and children. To compare the se- quences’ lengths and number of PNGs between family members, a non parametric Wilcoxon rank-sum (Mann- Whitney) test was used. Tests of statistical significance included the 95% confidence interval of relative risks and two sided p-values. Similar analysis was done after stratifying the study population by age and gender for both heterosexual and vertical transmission events. Indels assessment along the sequences was done manually. The 520 base pair nucleotide sequences were translated to amino acid sequence using the Gene Doc program. The amino acid sequences in their fasta formats were entered into a glycosylation an alysis site: Copyright © 2011 SciRes. AID  HIV-1 Env gp120 C2V5 Potential N-Linked Glycosylation Site(s) (PNGs) Variations and Amino Acid Length Polymorphisms among Infected Family Members Copyright © 2011 SciRes. AID 3 http://www.hiv.lanl.gov /content/hiv-db/GLYCOSITE/gly cosite where PNGs were marked and counted. Variations in the number and location of PNGs within the entire C2V5 region an d sub-regions w ere analysed. Median and range values of sequence lengths and PNGs were calcu- lated for each family member as previously described by others [14]. 4. Ethical Consideration The study was approved by the Medical Research Coun- cil of Zimbabwe (MRCZ) and the Ethical Review Com- mittee in Norway. Written consent to participate in the HIV-1 genetic research study was obtained from the par- ent(s) who also consented on behalf of their minors. Study participants were free to discontinue at any time without any prejudice. Parents also consented to have their blood samples and those of their children to be used for other future HIV related research studies. 5. Results 5.1. Characteristics of the Four Families Three of the four couples were in a monogamous mar- riage except for Mother 375 who was in a polygamous relationship. All parents had at least seven years in school and were of low economic status. Mean age of the moth- ers and their spouses were 34.8 years ± 3.2 years and 38 years ± 2 years, respectively. 5.2. HIV Infections Parents of the four families were HIV-1 positive at en- rolment but did not know when and how they got in- fected. Mode of HIV-1 acquisition was most likely het- erosexually as none mentioned any history of blood transfusion, drug abuse or homosexuality except for mother 366 who had a history of blood transfusion hence it could not be substantiated whether she got infected heterosexually or through blood transfusion. Index chil- dren of families 366, 375 and 567 and second siblings of families 205 and 567 were HIV-1 DNA PCR negative at delivery and six weeks postpartum but later got infected through breastfeeding. Index child 205 was HIV-1 DNA PCR negative at delivery. However, he did not turn up for his 6 week postpartum visit hence his time of infec- tion was not definite. 5.3. Global Analysis of Families’ C2V5 Sub-Re- gions Nucleotide Sequences Consistent feature of families sequence analysis showed the highest variation within the env gp120 V4 and V5 and interestingly also within the C3 sub-regions. Indels char- acterized the genetic diversity of these three sub-regions. In contrast amino acid substitutions constituted the main cause of variability within the constant domains C2, C4 and to a lesser extent V3 region, see Figure 1. Of note was a 12 base pair nucleotide in sertion observed in Fam- ily 366 Mother-Infant pair viral sequences that was ab- sent in the purported father`s HIV sequence. 5.4. Glycosylation Patterns and Amino Acid Lengths of the Variable Loops, V3-V5 Of the variable regions, V3 was relatively conserved re- gardless of family member ag e or sex with no insertions, deletions or shifts in PNGs that ch aracterized V4 and V5 domains. Families 205, 366 and 375 V3 amino acid lengths were all 35, see Tables 1-3. Contrary, most se- quences of 567 family members were of 33 amino acids in length; consequently they had the least number of PNGs. Despite having the optimal amino acids length within the V3 region, Family 375 had a just single PNGs, see Tables 1-6. Remarkably was the presence of the conserved GPGQ motif within the V3 crown of all family members’ viral sequences that was preserved following both heterosexual and vertical transmission events. On the contrary, 2/5 of Mother 366 HIV-1 clones had GPGR motif instead. V4 region showed extensive amino acid length polymor- phism and PNGs variation ranging from median (range) 28(24 - 35) and 3(2 - 6) respectively, see Tables 1 and 6. There were numerous deletions in this reg ion resulting in variants with different V4 region amino acid length poly- morphisms as shown by sequence lengths of fathers 205 and 375 including Mother 567 and to some extent Mother 366. The first two PNGs had more fixed locations in the distribution whilst the other two were randomly distrib- uted. Interestingly, Father 205 lost a PNGs resulting in his major HIV-1 variants having just 3 PNGs. Insertions within Mother 366 HIV-1 sequences resulted in her gaining one more PNGs. Of note was the loss of the first PNGs within the V4 region observed in 3/4 of the index children, see Figure 1. V5 region PNGs varied from me- dian (range) 2 (1 - 3) between families, with Family 205 HIV-1 variants having an average of 3 per family mem- ber whilst just one PNGs was a common observation with Family 567. 5.5. Glycosylation Patterns and Amino Acid Lengths of the Constant Regions 5.5.1. Heterosexual Transmission There were no significant gender differences with respect to amino acid lengths between fathers and mothers for the entire C2V5 length, 207 (199 - 215); 208 (198 - 216), p = 0.734. However, after stratifying by sub-regions, there was a general tendency for mothers of families 366 and 375 to have longer C3 region amino acid lengths 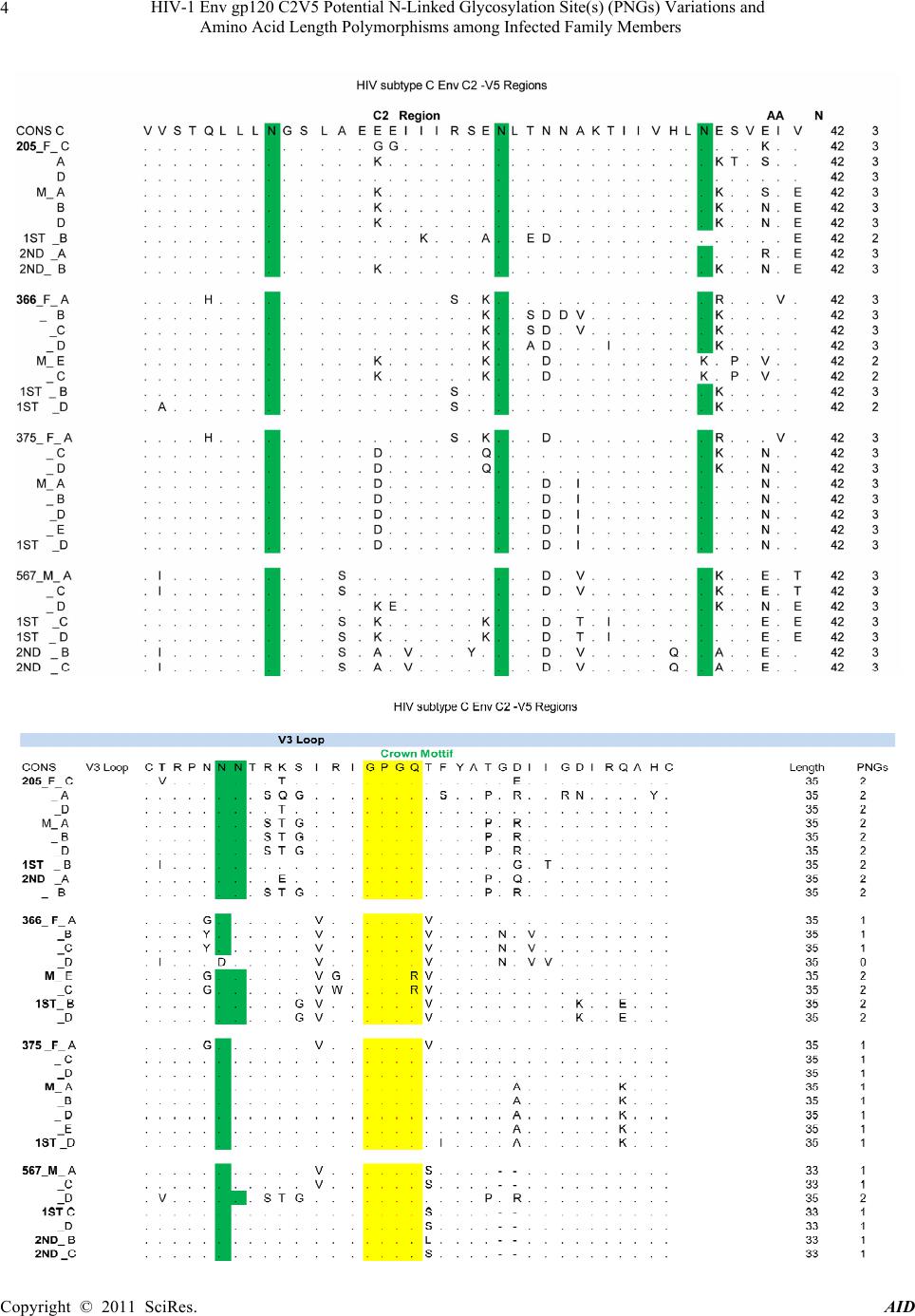 HIV-1 Env gp120 C2V5 Potential N-Linked Glycosylation Site(s) (PNGs) Variations and 4 Amino Acid Length Polymorphisms among Infected Family Members Copyright © 2011 SciRes. AID 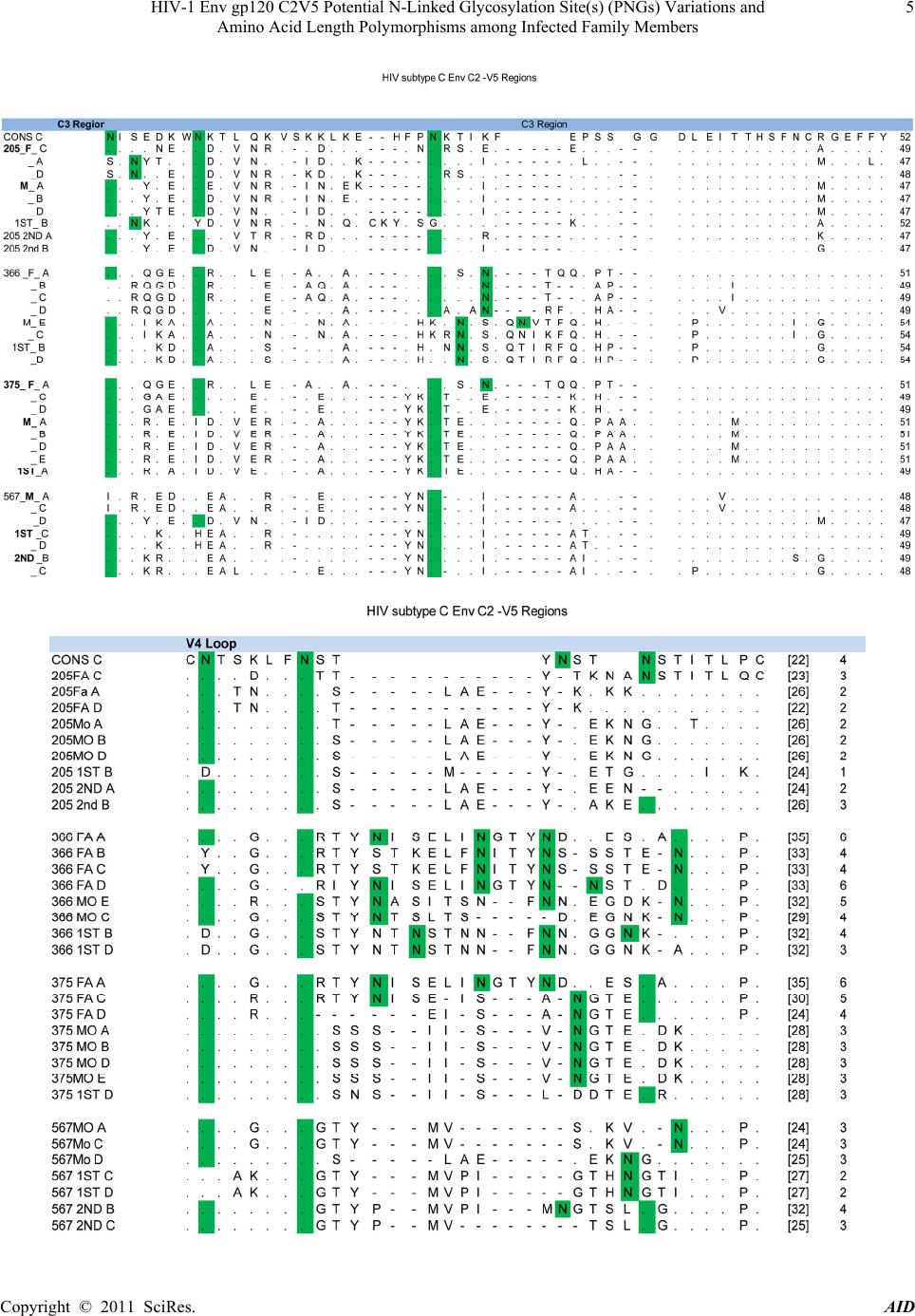 HIV-1 Env gp120 C2V5 Potential N-Linked Glycosylation Site(s) (PNGs) Variations and 5 Amino Acid Length Polymorphisms among Infected Family Members Copyright © 2011 SciRes. AID 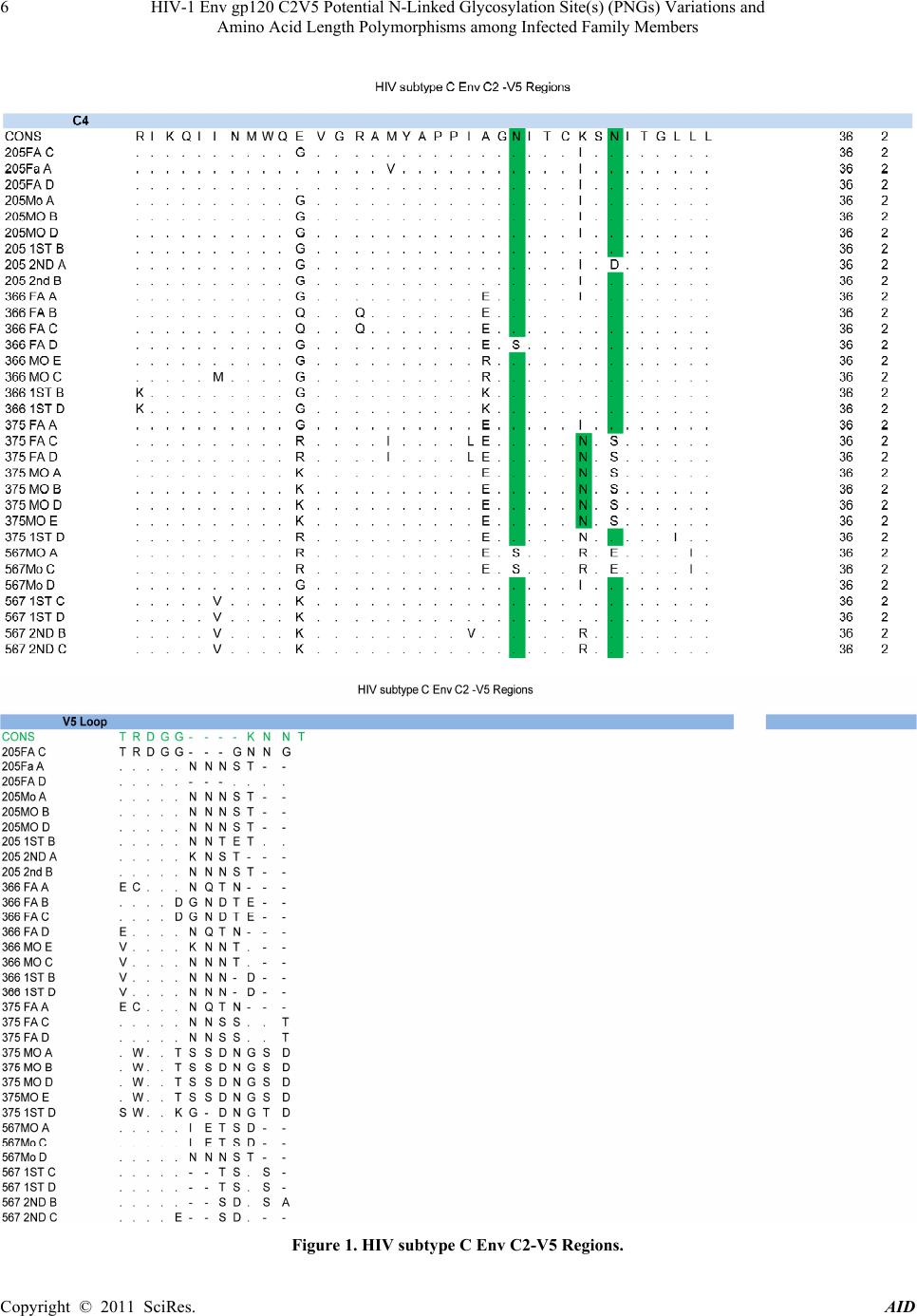 HIV-1 Env gp120 C2V5 Potential N-Linked Glycosylation Site(s) (PNGs) Variations and 6 Amino Acid Length Polymorphisms among Infected Family Members Figure 1. HIV subtype C Env C2-V5 Regions. Copyright © 2011 SciRes. AID 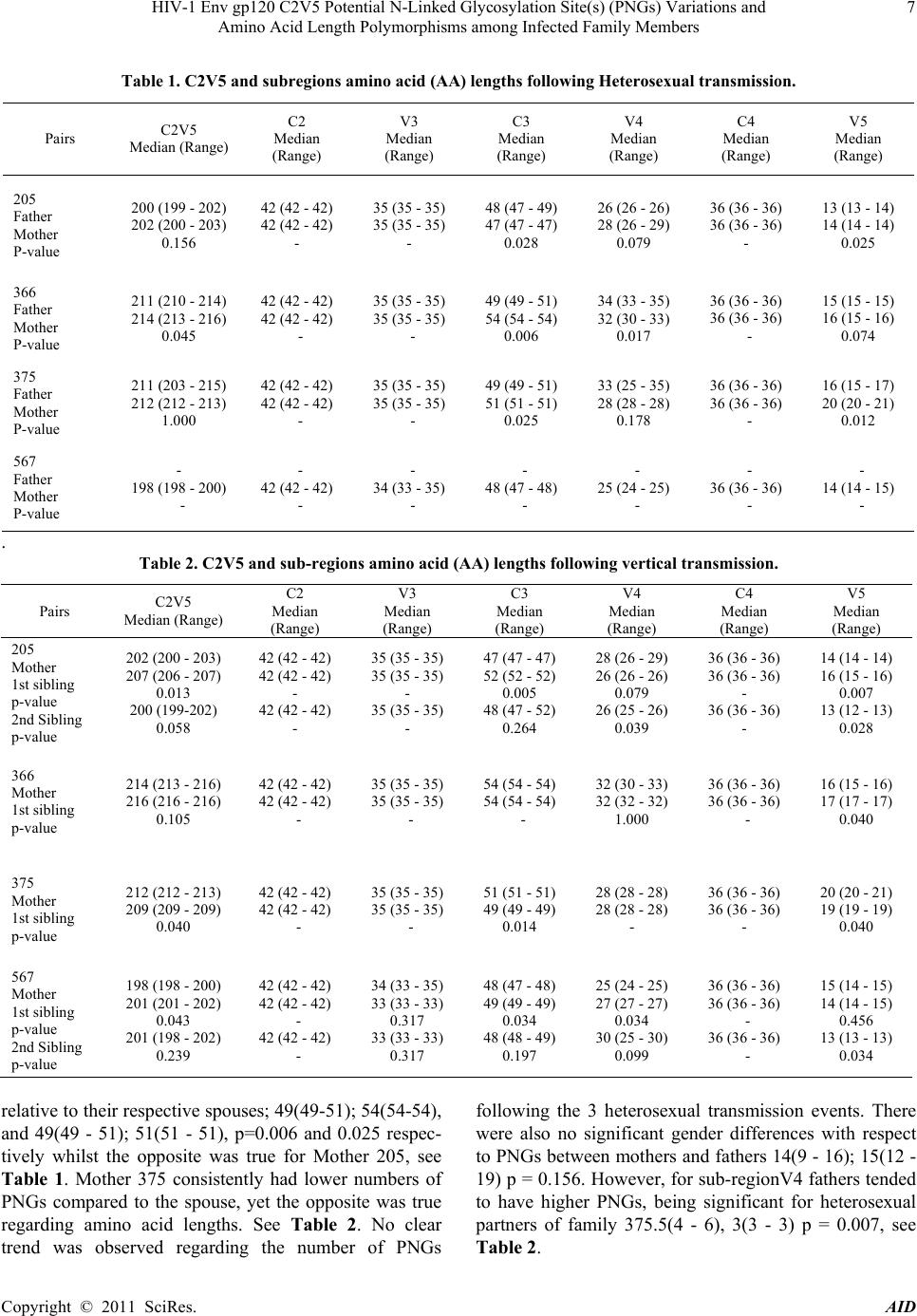 HIV-1 Env gp120 C2V5 Potential N-Linked Glycosylation Site(s) (PNGs) Variations and Amino Acid Length Polymorphisms among Infected Family Members Copyright © 2011 SciRes. AID 7 Table 1. C2V5 and subregions amino acid (AA) lengths following Heterosexual transmission. Pairs C2V5 Median (Range) C2 Median (Range) V3 Median (Range) C3 Median (Range) V4 Median (Range) C4 Median (Range) V5 Median (Range) 205 Father Mother P-value 200 (199 - 202) 202 (200 - 203) 0.156 42 (42 - 42) 42 (42 - 42) - 35 (35 - 35) 35 (35 - 35) - 48 (47 - 49) 47 (47 - 47) 0.028 26 (26 - 26) 28 (26 - 29) 0.079 36 (36 - 36) 36 (36 - 36) - 13 (13 - 14) 14 (14 - 14) 0.025 366 Father Mother P-value 211 (210 - 214) 214 (213 - 216) 0.045 42 (42 - 42) 42 (42 - 42) - 35 (35 - 35) 35 (35 - 35) - 49 (49 - 51) 54 (54 - 54) 0.006 34 (33 - 35) 32 (30 - 33) 0.017 36 (36 - 36) 36 (36 - 36) - 15 (15 - 15) 16 (15 - 16) 0.074 375 Father Mother P-value 211 (203 - 215) 212 (212 - 213) 1.000 42 (42 - 42) 42 (42 - 42) - 35 (35 - 35) 35 (35 - 35) - 49 (49 - 51) 51 (51 - 51) 0.025 33 (25 - 35) 28 (28 - 28) 0.178 36 (36 - 36) 36 (36 - 36) - 16 (15 - 17) 20 (20 - 21) 0.012 567 Father Mother P-value - 198 (198 - 200) - - 42 (42 - 42) - - 34 (33 - 35) - - 48 (47 - 48) - - 25 (24 - 25) - - 36 (36 - 36) - - 14 (14 - 15) - . Table 2. C2V5 and sub-regions amino acid (AA) lengths following vertical transmission. Pairs C2V5 Median (Range) C2 Median (Range) V3 Median (Range) C3 Median (Range) V4 Median (Range) C4 Median (Range) V5 Median (Range) 205 Mother 1st sibling p-value 2nd Sibling p-value 202 (200 - 203) 207 (206 - 207) 0.013 200 (199-202) 0.058 42 (42 - 42) 42 (42 - 42) - 42 (42 - 42) - 35 (35 - 35) 35 (35 - 35) - 35 (35 - 35) - 47 (47 - 47) 52 (52 - 52) 0.005 48 (47 - 52) 0.264 28 (26 - 29) 26 (26 - 26) 0.079 26 (25 - 26) 0.039 36 (36 - 36) 36 (36 - 36) - 36 (36 - 36) - 14 (14 - 14) 16 (15 - 16) 0.007 13 (12 - 13) 0.028 366 Mother 1st sibling p-value 214 (213 - 216) 216 (216 - 216) 0.105 42 (42 - 42) 42 (42 - 42) - 35 (35 - 35) 35 (35 - 35) - 54 (54 - 54) 54 (54 - 54) - 32 (30 - 33) 32 (32 - 32) 1.000 36 (36 - 36) 36 (36 - 36) - 16 (15 - 16) 17 (17 - 17) 0.040 375 Mother 1st sibling p-value 212 (212 - 213) 209 (209 - 209) 0.040 42 (42 - 42) 42 (42 - 42) - 35 (35 - 35) 35 (35 - 35) - 51 (51 - 51) 49 (49 - 49) 0.014 28 (28 - 28) 28 (28 - 28) - 36 (36 - 36) 36 (36 - 36) - 20 (20 - 21) 19 (19 - 19) 0.040 567 Mother 1st sibling p-value 2nd Sibling p-value 198 (198 - 200) 201 (201 - 202) 0.043 201 (198 - 202) 0.239 42 (42 - 42) 42 (42 - 42) - 42 (42 - 42) - 34 (33 - 35) 33 (33 - 33) 0.317 33 (33 - 33) 0.317 48 (47 - 48) 49 (49 - 49) 0.034 48 (48 - 49) 0.197 25 (24 - 25) 27 (27 - 27) 0.034 30 (25 - 30) 0.099 36 (36 - 36) 36 (36 - 36) - 36 (36 - 36) - 15 (14 - 15) 14 (14 - 15) 0.456 13 (13 - 13) 0.034 relative to their respective spou ses; 49(49-51); 54(54-54), and 49(49 - 51); 51(51 - 51), p=0.006 and 0.025 respec- tively whilst the opposite was true for Mother 205, see Table 1. Mother 375 consistently had lower numbers of PNGs compared to the spouse, yet the opposite was true regarding amino acid lengths. See Table 2. No clear trend was observed regarding the number of PNGs following the 3 heterosexual transmission events. There were also no significant gender differences with respect to PNGs between mothers and fathers 14(9 - 16); 15(12 - 19) p = 0.156. Howeve r, for sub-regionV4 fathers tend ed to have higher PNGs, being significant for heterosexual partners of family 375.5(4 - 6), 3(3 - 3) p = 0.007, see Table 2.  HIV-1 Env gp120 C2V5 Potential N-Linked Glycosylation Site(s) (PNGs) Variations and 8 Amino Acid Length Polymorphisms among Infected Family Members Table 3. C2V5 and sub-regions amino acid (AA) lengths variations following PTC transmission. Pairs C2V5 Median (Range) C2 Median (Range) V3 Median (Range) C3 Median (Range) V4 Median (Range) C4 Median (Range) V5 Median (Range) 205 Father 1st sibling p-value 2nd Sibling p-value 200 (199 - 202) 207 (206 - 207) 0.019 200 (199 - 202) 0.350 42 (42 - 42) 42 (42 - 42) - 42 (42 - 42) - 35 (35 - 35) 35 (35 - 35) - 35 (35 - 35) - 48 (47 - 49) 52 (52 - 52) 0.013 48 (47 - 52) 0.439 26 (26 - 26) 26 (26 - 26) - 26 (25 - 26) 0.127 36 (36 - 36) 36 (36 - 36) - 36 (36 - 36) - 13 (13 - 14) 16 (15 - 16) 0.017 13 (12 - 13) 0.617 366 Father 1st sibling p-value 211 (210 - 214) 216 (216 - 216) 0.057 42 (42 - 42) 42 (42 - 42) - 35 (35 - 35) 35 (35 - 35) - 49 (49 - 51) 54 (54 - 54) 0.046 34 (33 - 35) 32 (32 - 32) 0.057 36 (36 - 36) 36 (36 - 36) - 15 (15 - 15) 17 (17 - 17) 0.025 375 Father 1st sibling p-value 211 (203 - 215) 209 (209 - 209) 1.000 42 (42 - 42) 42 (42 - 42) - 35 (35 - 35) 35 (35 - 35) - 49 (49 - 51) 49 (49 - 49) 0.480 33 (25 - 35) 28 (28 - 28) 0.340 36 (36 - 36) 36 (36 - 36) - 16 (15 - 17) 19 (19 - 19) 0.057 567 Father 1st sibling p-value 2nd Sibling p-value - 201 (201 - 202) 0.043 201 (198 - 202) 0.239 - 42 (42 - 42) - 42 (42 - 42) - - 33 (33 - 33) 0.317 33 (33 - 33) 0.317 - 49 (49 - 49) 0.034 48 (48 - 49) 0.197 - 27 (27 - 27) 0.034 30 (25 - 30) 0.099 - 36 (36 - 36) - 36 (36 - 36) - - 14 (14 - 15) 0.456 13 (13 - 13) 0.034 Table 4. Heterosexual patterns following transmission (PNGS). Pairs C2V5 Median (Range) C2 Median (Range) V3 Median (Range) C3 Median (Range) V4 Median (Range) C4 Median (Range) V5 Median (Range) 205 Father Mother p-value 13 (12 - 15) 16 (15 - 16) 0.014 3 (3 - 3) 3 (3 - 3) - 2 (1 - 2) 2 (1 - 2) 0.371 3 (2 - 3) 3 (3 - 3) 0.091 3 (3 - 3) 3 (3 - 3) - 2 (2 - 2) 2 (2 - 2) - 1 (1 - 2) 3 (3 - 3) 0.006 366 Father Mother p-value 16 (14 - 19) 15 (14 - 16) 0.379 3 (3 - 3) 2 (2 - 2) 0.005 1 (0 - 1) 2 (2 - 2) 0.006 3 (3 - 4) 3 (3 - 4) 0.866 5 (4 - 6) 4 (3 - 5) 0.302 2 (1 - 2) 2 (2 - 2) 0.264 3 (2 - 3) 2 (1 - 2) 0.058 375 Father Mother p-value 16 (15 - 18) 13 (13 - 13) 0.007 3 (3 - 3) 3 (3 - 3) - 1 (1 - 1) 1 (1 - 1) - 3 (3 - 4) 2 (2 - 2) 0.006 5 (4 - 6) 3 (3 - 3) 0.007 2 (2 - 2) 2 (2 - 2) - 2 (2 - 2) 2 (2 - 2) - 567 Father Mother p-value - 10 (9 - 11) - - 3 (3 - 3) - - 1 (1 - 2) - - 1 (1 - 3) - - 3 (3 - 3) - - 0 (0 - 0) - - 1 (1 - 1) - C2 and C4 regions sequence lengths were exactly the same for all families‟ heterosexual partners but differed with respect to C3, V4 and V5 sub-regions. Applying the postulated hypothesis to C3 and to a lesser extent V5 sub-region sequence lengths [14], it is tantamount to tempting to propose the direction of heterosexual trans- mission, a very sensitive issue and should be handled carefully. A male to female (MTF) heterosexual trans- mission was suspected for family 205 with the father probably infecting his wife whilst female to male (FTM) transmission events were possible for families 366 and 375. Interestingly, though not statistically significant, an exact opposite trend was observed for the V4 variable region, see Table 1. 5.5.2. PTCT and Sequence Lengths and PNGs There were no significant V4 region amino acid lengths differences with respect to the C2, V3 and to some extent C4 sub-regions between mothers and first siblings. However, for the entire C2V5 region there was a tendency Copyright © 2011 SciRes. AID  HIV-1 Env gp120 C2V5 Potential N-Linked Glycosylation Site(s) (PNGs) Variations and 9 Amino Acid Length Polymorphisms among Infected Family Members Table 5. PNG patterns following transmission. Pairs C2V5 Median (Range) C2 Median (Range) V3 Median (Range) C3 Median (Range) V4 Median (Range) C4 Median (Range) V5 Median (Range) 205 Mother 1st sibling p-value 2nd Sibling p-value 16 (15 - 16) 11 (10 - 13) 0.010 14 (12 - 16) 0.080 3 (3 - 3) 2 (2 - 2) 0.005 3 (3 - 3) - 2 (1 - 2) 1 (1 - 2) 0.120 2 (1 - 2) 0.866 3 (3 - 3) 2 (1 - 2) 0.007 3 (1 - 3) 0.264 3 (3 - 3) 3 (3 - 3) - 3 (2 - 3) 0.264 2 (2 - 2) 2 (2 - 2) - 2 (1 - 2) 0.264 3 (3 - 3) 2 (1 - 2) 0.007 2 (2 - 3) 0.025 366 Mother 1st sibling p-value 15 (14 - 16) 14 (13 - 15) 0.320 2 (2 - 2) 3 (2 - 3) 0.114 2 (2 - 2) 2 (2 - 2) - 3 (3 - 4) 3 (3 - 3) 0.527 4 (3 - 5) 4 (3 - 4) 0.306 2 (2 - 2) 2 (2 - 2) - 2 (1 - 2) 1 (1 - 1) 0.180 375 Mother 1st sibling p-value 13 (13 - 13) 12 (11 - 13) 0.114 3 (3 - 3) 3 (3 - 3) - 1 (1 - 1) 1 (1 - 1) - 2 (2 - 2) 2 (2 - 2) - 3 (3 - 3) 3 (3 - 3) - 2 (2 - 2) 1 (0 - 2) 0.114 2 (2 - 2) 2 (2 - 2) - 567 Mother 1st sibling p-value 2nd Sibling p-value 10 (9 - 11) 12 (12 - 12) 0.037 13 (12 - 13) 0.046 3 (3 - 3) 3 (3 - 3) - 3 (3 - 3) - 1 (1 - 2) 1 (1 - 1) 0.317 1 (1 - 1) 0.317 1 (1 - 3) 2 (2 - 2) 0.480 2 (2 - 2) 0.480 3 (3 - 3) 2 (2 - 2) 0.025 4 (3 - 4) 0.114 0 (0 - 0) 2 (2 - 2) 0.025 2 (2 - 2) 0.025 1 (1 - 1) 2 (2 - 2) 0.025 1 (1 - 1) - Table 6. PNG patterns following PTC transmission. Pairs C2V5 Median (Range) C2 Median (Range) V3 Median (Range) C3 Median (Range) V4 Median (Range) C4 Median (Range) V5 Median (Range) 205 Father 1st sibling p-value 2nd Sibling p-value 13 (12 - 15) 11 (10 - 13) 0.080 14 (12 - 16) 0.457 3 (3 - 3) 2 (2 - 2) 0.008 3 (3 - 3) - 2 (1 - 2) 1 (1 - 2) 0.495 2 (1 - 2) 0.495 3 (2 - 3) 2 (1 - 2) 0.061 3 (1 - 3) 0.739 3 (3 - 3) 3 (3 - 3) - 3 (2 - 3) 0.317 2 (2 - 2) 2 (2 - 2) - 2 (1 - 2) 0.317 1 (1 - 2) 2 (1 - 2) 0.495 2 (2 - 3) 0.040 366 Father 1st sibling p-value 16 (14 - 19) 14 (13 - 15) 0.240 3 (3 - 3) 3 (2 - 3) 0.157 1 (0 - 1) 2 (2 - 2) 0.046 3 (3 - 4) 3 (3 - 3) 0.480 5 (4 - 6) 4 (3 - 4) 0.134 2 (1 - 2) 2 (2 - 2) 0.480 3 (2 - 3) 1 (1 - 1) 0.053 375 Father 1st sibling p-value 16 (15 - 18) 12 (11 - 13) 0.060 3 (3 - 3) 3 (3 - 3) - 1 (1 - 1) 1 (1 - 1) - 3 (3 - 4) 2 (2 - 2) 0.046 5 (4 - 6) 3 (3 - 3) 0.057 2 (2 - 2) 1 (0 - 2) 0.157 2 (2 - 2) 2 (2 - 2) - 567 Father 1st sibling p-value 2nd Sibling p-value - 12 (12 - 12) 0.037 13 (12 - 13) 0.046 - 3 (3 - 3) - 3 (3 - 3) - - 1 (1 - 1) 0.317 1 (1 - 1) 0.317 - 2 (2 - 2) 0.480 2 (2 - 2) 0.480 - 2 (2 - 2) 0.025 4 (3 - 4) 0.114 - 2 (2 - 2) 0.025 2 (2 - 2) 0.025 - 2 (2 - 2) 0.025 1 (1 - 1) - Copyright © 2011 SciRes. AID  HIV-1 Env gp120 C2V5 Potential N-Linked Glycosylation Site(s) (PNGs) Variations and 10 Amino Acid Length Polymorphisms among Infected Family Members for the index children to have longer amino acid lengths compared to the mothers p = 0.013, 0.040 and 0.043 for families 205, 375 an d 567 respectively, see Table 2. Dif- ferences were also observed within the V5 sub-region with p values =0.007, 0.040, 0.040 and 0.456 for Fami- lies 205, 366, 375 and 567 respectively, but without any clear trend. Family 205 second siblings’ V4 and V5 sub-regions amino acid lengths were both significantly shorter relative to the maternal ones; p=0.039 and 0.028 respectively. Similar observations were also noted for 567 second sibling sequences which were significantly shorter relative to maternal ones p=0.034. Significant differences were also observed in the C3 region but with no clear trends. See Table 2. Interestingly a similar trend like the one observed in the mothers with first siblings was also observed with paternal sequences with respect to the C2V5, C3, and V5 regions. However, there were no differences in sequence lengths between paternal ones and those from second sibling, see Table 3. There was a tendency for parents to have longer amino acid lengths for the regions C2V5 and V3 being; 208(198 - 216); 205 (198 - 216): p = 0.138 and 35(33 - 35); 34(33 - 35): p = 0.018. Following stratification by age, there was also a tendency for parents to have denser glycans within the C2V5, C3 and V4 reg ions which were 14(9 - 19); 13 (10 - 16), 3(1 - 4); 2(1 - 3) and 4(3 - 6); 3(2 - 4) p= 0.0025, 0.005 and 0.008 respectively. For both the entir e C2V5 region and sub-region C3 the first siblings had significantly longer amino acid lengths relative to the second sib lings; 207(201 - 216), 200(198 - 202) and 51(49 - 54), 48(47 - 52); p = 0.005 and 0.007 respectively. However, the opposite was true for the V5 region where the second siblings had much longer amino acid lengths 13(12 - 14), 11(14 - 19); p= 0.0006. After gender stratification no differences were observed re- garding amino acid lengths whilst female siblings tended to have a denser glycan shield relative to their male counterparts, 14(11 - 16); 12 (10 - 13); p = 0.039. Gly- cosylation patterns found in siblings had similar patterns to those found in the parents, see Table 6. 6. Discussion N-glycosylation is an essential co-translational modifica- tion process where a sugar, glycan, is covalently attached to the amide group of asparagine (N) residues [29].The N-linked glycosylation of the HIV-1 env gp120 is one of the most important viral protective mechanisms to over- come in order to develop an effective neutralizing anti- body-inducing vaccine. Since several PNGs are relatively constant across HIV-1 subtypes, there is a great deal of interest in developing a carbohydrate based antigen de- signed to elicit a humoral immune response. Studies have shown a subtype env sequence length dependency fol- lowing transmission with subtypes A and C significantly shifted towards shor ter lengths though untrue for subtype B [14, 20, 30]. Our results are similar to previous subtype A and D heterosexual transmission where trends were observed for HIV-1 env lengths but no significant differ- ences were observed with PNGs [19]. For PTCT, our study observed children being infected with shorter sequences demonstrated by significantly shorter sequences in the second sibling relative to the maternal or paternal sequences which tended to increase with age or disease progression, probably due to immune selection as shown in the first siblings who had signifi- cantly longer sequ ences and PNGs relative to the pa rents. Our results are suggestive of the fact that a child acquires more or less the same number of PNGs like the mother and these tend to decrease with time or disease progres- sion as evidenced by the presence of the first V4 region PNGs observed amongst the second siblings who were about 14 months old but was absent or lost in 3 out of 4 of the first siblings who were about five years old. This observation may have something to do with functional elements of V4 region which exist solely to facilitate viral escape during the evolving glycan shield [41]. PNGs number differences were also observed in chronic HIV-1 infection [31-33]. Similar to our findings some pediatric HIV-1 env subtype C studies have also demonstrated no clear pattern of change in the number PNGs [34,35]. Mother 567 with the least number of PNGs has since died during beginning of this year suggestive of the fact that loss of PNGs may be associated with advanced disease and probably a risk factor of dying. It would be interest- ing to correlate PNGs with markers of disease progres- sion. A consistent feature of families’ sequence analysis showed the highest levels of variation within the env gp120, V4 and V5 variable including the C3 regions challenging whether the so-called “constant region” with respect to subtype B is really also a constant one when it comes to subtype C. More so, atypically, our families’ HIV-1 subtype C V3 region was relatively constant, again querying the currently common practice of ex- trapolating subtype B findings to non-subtype B ones. Traditionally, V3 region has been considered a variable domain based on analysis of subtype B sequences. How- ever, the small numbers of clones sequenced may not have been sufficient for statistically sound conclusions. A larger sample size is recommended to substantiate these findings. Interestingly, similar results of a relatively con- stant subtype C V3 region have been obtained elsewhere [36-39]. Conserved PNGs within the V3 region amongst all family members regardless of age or sex points to Copyright © 2011 SciRes. AID  HIV-1 Env gp120 C2V5 Potential N-Linked Glycosylation Site(s) (PNGs) Variations and 11 Amino Acid Length Polymorphisms among Infected Family Members their potentially important functional elements which may function primarily to mask the large number of neu- tralizing antibody ep itopes defined in this region. As pre- viously reported by others, emergence of deletions within the V4 region was also observed among all family mem- bers both adults and children [40]. These coupled with amino acid substitutions affected the number and distri- bution of PNGs resulting in the coexistence within some family members V4 variants each characterized by dif- ferent amino acid sequences and PNGs patterns. Previous subtype C studies have showed extensive V4 length polymorphism where shorter lengths have been shown to enhance infectivity at the cost of exposing neutralizing epitopes [41]. Some countries including Zimbabwe now seek to in- stitute legislation to hold persons criminally responsible for willfully infecting others with HIV. This law has been unsuccessfully applied as it is very difficult to prove criminal intent on the part of the alleged transmitter and currently there is no supporting forensic evidence to prove directionality of infection. For successful prosecu- tion of willful transmission, ascertaining the association of number of PNGs or the HIV-1 env domain length polymorphism and age of infection may be the first step to provide a line of eviden ce to that effect, hence the need for bigger studies. 7. Conclusions Highest env gp120 region variation characterised by in- dels were observed within theC3, V4 and V5 sub-regions. Indels character ized s. Our study su ppor ts th e ob serv ation that HIV-1 env C2V5 sequences and PNGs tend to in- crease with age and disease progression in children. Though sensitive and should be handled carefully, with respect to C3 and V5 sequence lengths it is tempting to propose a possible male to female and female to male heterosexual transmission events for families 205 and 366 including 375 respectively. Ascertaining the associa- tion of PNGs or env sequence length polymorphism and period of infection may be the first step towards a possi- ble line of forensic evidence. Hence bigger studies are warranted to substantiate the authenticity of this poten- tially useful application. 8. Acknowledgements Acknowledgements are due to Drs. Kurewa NE and Munjoma MW including all the support staff for facili- tating sample collection. Many thanks are due to the families who participated in this study. Special mention goes to the Letten Foundation and Professor Letten F Saugstad herself for funding the study. REFERENCES [1] B. R. Starcich, B. H. Hahn, G. M. Shaw, P. D. McNeely, S. Modrow, H. Wolf, et al., “Identification and Charac- terization of Conserved and Variable Regions in the En- velope Gene of HTLV-III/LAV, the Retrovirus of AIDS,” Cell, Vol. 45, No. 5, 1986, pp. 637-648 [2] H. Geyer, C. Holschbach, G. Hunsmann and J. Schneider, “Carbohydrates of Human Immunodeficiency Virus. Structures of Oligosaccharides Linked to the Envelope Glycoprotein 120,” The Journal of Biological Chemistry, Vol. 263, No. 24, 1988, pp. 11760-11767. [3] M. Zhang, B. Gaschen, W. Blay, B. Foley, N. Haigwood, C. Kuiken, et al., “Tracking Global Patterns of N-Linked Glycosylation Site Variation in Highly Variable Viral Glycoproteins: HIV, SIV, and HCV Envelopes and influ- enza Hemagglutinin,” Glycobiology, Vol. 14, No. 12, 2004, pp. 1229-1246. [4] C. K. Leonard, M. W. Spellman, L. Riddle, R. J. Harris, J. N. Thomas and T. J. Gregory, “Assignment of Intrachain Disulfide Bonds and Characterization of Potential Glyco- sylation Sites of the Type 1 Recombinant Human Immu- nodeficiency Virus Envelope Glycoprotein (gp120) Ex- pressed in Chinese Hamster Ovary Cells,” The Journal of Biological Chemistry, Vol. 265, No. 18, 1990, pp. 10373-10382. [5] P. D. Kwong, R. Wyatt, Q. J. Sattentau, J. Sodroski, W. A. Hendrickson, “Oligomeric Modeling and Electrostatic Analysis of the gp120 Envelope Glycoprotein of Human Immunodeficiency Virus,” The Journal of Virology, Vol. 74, No. 4, 2000, pp. 1961-1972. doi:10.1128/JVI.74.4.1961-1972.2000 [6] R. Wyatt, P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, et al., “The Antigenic Structure of the HIV gp120 Envelope Glycoprotein,” Na- ture, 1998, Vol. 393, No. 6686, pp. 705-711. [7] J. J. Skehel, D. J. Stevens, R. S. Daniels, A. R. Douglas, M. Knossow, I. A. Wilson, et al., “A Carbohydrate Side Chain on Hemagglutinins of Hong Kong Influenza Vi- ruses Inhibits Recognition by a Monoclonal Antibody,” Proceedings of the National Academy of Sciences, Vol. 81, No. 6, 1984, pp. 1779-1783. doi:10.1073/pnas.81.6.1779 [8] P. Botarelli, B. A. Houlden, N. L. Haigwood, C. Servis, D. Montagna and S. Abrignani, “N-Glycosylation of HIV- gp120 May Constrain Recognition by T Lymphocytes,” The Journal of Immunology, Vol. 147, No. 9, 1991, pp. 3128-3132. [9] Cheng-Mayer C., Brown A., Harouse J., Luciw P.A. and Mayer A.J., “Selection for Neutralization Resistance of the Simian/Human Immunodeficiency Virus SHIVSF33A Variant in vivo by Virtue of Sequence Changes in the Ex- tracellular Envelope Glycoprotein that Modify N-Linked Glycosylation,” The Journal of Virology, Vol. 73, No. 7, 1999, pp. 5294-5300. [10] X. Wei, J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, et al., “Antibody Neutralization and Escape by HIV-1,” Nature, Vol. 422, No. 6929, 2003, pp. 307-312. Copyright © 2011 SciRes. AID  HIV-1 Env gp120 C2V5 Potential N-Linked Glycosylation Site(s) (PNGs) Variations and 12 Amino Acid Length Polymorphisms among Infected Family Members doi:10.1128/JVI.00141-06 [11] M. Sagar, X. Wu, S. Lee and J. Overbaugh, “Human Im- munodeficiency Virus Type 1 V1-V2 Envelope Loop Se- quences Expand and Add Glycosylation Sites over the Course of Infection, and These Modifications Affect An- tibody Neutralization Sensitivity,” The Journal of Virol- ogy, 2006, Vol. 80, No. 19, pp. 9586-9598. [12] E. M. Bunnik, L. Pisas, A. C. van Nuenen and H. Schuitemaker, “Autologous Neutralizing Humoral Immu- nity and Evolution of the Viral Envelope in the Course of Subtype B Human Immunodeficiency Virus Ty pe 1 Infec- tion,” The Journal of Virology, Vol. 82 No. 16, 2008, pp. 7932-7941. doi:10.1128/JVI.00757-08 [13] H. F. Gunthard, S. D. Frost, A. J. Leigh-Brown, C. C. Ignacio, K. Kee, A. S. Perelson, et al., “Evolution of En- velope Sequences of Human Immunodeficiency Virus Type 1 in Cellular Reservoirs in the Setting of Potent An- tiviral Therapy,” The Journal of Virology, 1999, Vol. 73, No. 11, pp. 9404-9412 [14] C. A. Derdeyn, J. M. Decker, F. Bibollet-Ruche, J. L. Mokili, M. Muldoon, S. A. Denham, et al., “Enve- lope-Constrained Neutralization-Sensitive HIV-1 after Heterosexual Transmission,” Science, 2004, Vol. 303, No. 5666, pp. 2019-2022. [15] B. Li, J.M. Decker, R. W. Johnson, F. Bibollet-Ruche, X. Wei, J. Mulenga, et al., “Evidence for Potent Autologous Neutralizing Antibody Titers and Compact Envelopes in Early Infection with Subtype C Human Immunodefi- ciency Virus Type 1,” The Journal of Virology, Vol. 80, No. 11, 2006, pp. 5211-5218. doi:10.1128/JVI.00201-06 [16] J. Irungu, E. P. Go, Y. Zhang, D. S. Dalpathado, H. X. Liao, B. F. Haynes, et al., “Comparison of HPLC/ESI- FTICR MS versus MALDI-TOF/TOF MS for Glycopep- tide Analysis of a Highly Glycosylated HIV Envelope Glycoprotein,” Journal of the American Society for Mass Spectrometry, Vol. 19, No. 8, 2008, pp. 1209-1220. doi:10.1016/j.jasms.2008.05.010 [17] H. Zhang, D. C. Tully, F. G. Hoffmann, J. He, C. Kankasa and C. Wood, “Restricted Genetic Diversity of HIV-1 Subtype C Envelope Glycoprotein from Perinatally In- fected Zambian Infants,” PLoS One, Vol. 5, No. 2, 2010, p. e9294. doi:10.1371/journal.pone.0009294 [18] M. Alexander, R. Lynch, J. Mulenga, S. Allen, C. A. Derdeyn and E. Hunter, “Donor and Recipient Envs from Heterosexual Human Immunodeficiency Virus Subtype C Transmission Pairs Require High Receptor Levels for En- try,” The Journal of Virology, Vol. 84, No. 8, 2010, pp. 4100-4104. doi:10.1128/JVI.02068-09 [19] M. Sagar, O. Laeyendecker, S. Lee, J. Gamiel, M. J. Wawer, R. H. Gray, et al., “Selection of HIV Variants with Signature Genotypic Characteristics during Hetero- sexual Transmission,” Journal of Infectious Diseases, Vol. 199, No. 4, 2009, pp. 580-589. doi:10.1086/596557 [20] Y. Liu, M. E. Curlin, K. Diem, H. Zhao, A. K. Ghosh, H. Zhu, et al., “Env Length and N-Linked Glycosylation Following Transmission of Human Immunodeficiency Virus Type 1 Subty pe B Viruses,” Virology, Vol. 374, No. 2, 2008, pp. 229-233 [21] B. Chohan, D. Lang, M. Sagar, B. Korber, L. Lavreys, B. Richardson, et al., “Selection for Human Immunodefi- ciency Virus Type 1 Envelope Glycosylation Variants with Shorter V1-V2 Loop Sequences Occurs during Transmission of Certain Genetic Subtypes and May Im- pact Viral RNA Levels,” The Journal of Virology, 2005, Vol. 79, No. 10, pp. 6528-6531. doi:10.1128/JVI.79.10.6528-6531.2005 [22] T. Dieltjens, N. Loots, K. Vereecken, K. Grupping, L. Heyndrickx, E. Bottieau, et al., “HIV Type 1 Subtype A Envelope Genetic Evolution in a Slow Progressing Indi- vidual with Consistent Broadly Neutralizing Antibodies,” AIDS Research and Human Retroviruses, Vol. 25, No. 11, 2009, pp. 1165-1169. doi:10.1089/aid.2008.0161 [23] Y. Ye, Z. H. Si, J. P. Moore and J. Sodroski, “Association of Structural Changes in the V2 and V3 Loops of the gp120 Envelope Glycoprotein with Acquisition of Neu- tralization Resistance in a Simian-Human Immunodefi- ciency Virus Passaged in vivo,” The Journal of Virology, Vol. 74, No. 24, 2000, pp. 11955-11962. doi:10.1128/JVI.74.24.11955-11962.2000 [24] Y. Li, M. A. Rey-Cuille and S. L. Hu, “N-Linked Glyco- sylation in the V3 Region of HIV Type 1 Surface Antigen Modulates Coreceptor Usage in Viral Infection,” AIDS Research and Human Retroviruses, Vol. 17, No. 16, 2001, pp. 1473-1479. doi:10.1089/08892220152644179 [25] R. M. Lynch, T. Shen, S. Gnanakaran and C. A. Derdeyn, “Appreciating HIV Type 1 Diversity: Subtype Differences in Env,” AIDS Research and Human Retroviruses, Vol. 25, No. 3, 2009, pp. 237-248. doi:10.1089/aid.2008.0219 [26] C. N. Scanlan, J. Offer, N. Zitzmann and R. A. Dwek, “Exploiting the Defensive Sugars of HIV- 1 for Drug and Vaccine Design,” Nature, Vol. 446, No. 7139, 2007, pp. 1038-1045. [27] F. Z. Gumbo, G. Q. Kandawasvika, K. Duri, M. P. Map- ingure, N. E. Kurewa, K. Nathoo, et al., “Reduced HIV Transmission at Subsequent Pregnancy in a Re- source-Poor Setting,” Tropical Doctor, Vol. 41, No. 3, 2011, pp. 132-135. [28] K. Duri, W. Soko, F. Gumbo, K. Kristiansen, M. Mapin- gure, B. Stray-Pedersen, et al., “Genotypic Analysis of Human Immunodeficiency Virus Type 1 Env V3 Loop Sequences: Bioinformatics Prediction of Coreceptor Us- age among 28 Infected Mother-Infant Pairs in a Drug-Naive Population,” AIDS Research and Human Retroviruses, Vol. 27, No. 4, 2011, pp. 411-419. doi:10.1089/aid.2010.0142 [29] R. G. Spiro, “Protein glycosylation: Nature, Distribution, Enzymatic Formation, and Disease Implications of Gly- copeptide Bonds,” Glycobiology, Vol. 12, No. 4, 2002, pp. 43R-56R. [30] S. Gnanakaran, D. Lang, M. Daniels, T. Bhattacharya, C. A. Derdeyn and B. Korber, “Clade-Specific Differences between Human Immunodeficiency Virus Type 1 Clades B and C: Diversity and Correlations in C3-V4 Regions of gp120,” The Journal of Virology, Vol. 81, No. 9, 2007, pp. Copyright © 2011 SciRes. AID  HIV-1 Env gp120 C2V5 Potential N-Linked Glycosylation Site(s) (PNGs) Variations and Amino Acid Length Polymorphisms among Infected Family Members Copyright © 2011 SciRes. AID 13 4886-4891. doi:10.1128/JVI.01954-06 [31] N. Chomont, H. Hocini, G. Gresenguet, C. Brochier, H. Bouhlal, L. Andreoletti, et al., “Early Archives of Ge- netically-Restricted Proviral DNA in the Female Genital Tract after Heterosexual Transmission of HIV-1,” AIDS, Vol. 21, No. 2, 2007, pp. 153-162. [32] K.S. Kemal, B. Foley, H. Burger, K. Anastos, H. Minkoff, C. Kitchen, et al., “HIV-1 in Genital Tract and Plasma of Women: Compartmentalization of Viral Sequences, Core- ceptor Usage, and Glycosylation,” Proceedings of the Na- tional Academy of Sciences , Vol. 100, No. 22, 2003, pp. 12972-12977. doi:10.1073/pnas.2134064100 [33] S. K. Pillai, B. Good, S. K. Pond, J. K. Wong, M. C. Strain, D. D. Richman, et al., “Semen-Specific Genetic Characteristics of Human Immunodeficiency Virus Type 1 Env,” The Journal of Virology, Vol. 79, No. 3, 2005, pp. 1734-1742. doi:10.1128/JVI.79.3.1734-1742.2005 [34] F. Y. Tso, F.G. Hoffmann, D. C. Tully, P. Lemey, R. A. Rasmussen, H. Zhang, et al., “A Comparative Study of HIV-1 Clade C Env Evolution in a Zambian Infant with an Infected Rhesus Macaque during Disease Progression,” AIDS, Vol. 23, No. 14, 2009, pp. 1817-1828. [35] H. Zhang, F. Hoffmann, J. He, X. He and C. Kankasa, J. T. West, et al., “Characterization of HIV-1 Subtype C envelope Glycoproteins from Perinatally Infected Chil- dren with Different Courses of Disease,” Retrovirology, Vol. 3, 2006, p. 73. [36] B. T. Korber, K. MacInnes, R. F. Smith and G. Myers, “Mutational Trends in V3 Loop Protein Sequences Ob- served in Different Genetic Lineages of Human Immuno- deficiency Virus Type 1,” The Journal of Virology, Vol. 68, No. 10, 1994, pp. 6730-6744. [37] M. B. Patel, N. G. Hoffman and R. Swanstrom, “Sub- type-Specific Conformational Differences within the V3 Region of Subtype B and Subtype C Human Immunode- ficiency Virus Type 1 Env Proteins,” The Journal of Vi- rology, Vol. 82, No. 2, 2008, pp. 903-916. doi:10.1128/JVI.01444-07 [38] R. L. Stanfield, M. K. Gorny, S. Zolla-Pazner and I. A. Wilson, “Crystal Structures of Human Immunodeficiency Virus Type 1 (HIV-1) Neutralizing Antibody 2219 in Complex with Three Different V3 Peptides Reveal a New Binding Mode for HIV-1 Cross-Reactivity,” The Journal of Virology, Vol. 80, No. 12, 2006, pp. 6093-6105. doi:10.1128/JVI.00205-06 [39] P. B. Gilbert, V. Novitsky and M. Essex, “Covariability of Selected Amino Acid Positions for HIV Type 1 Subtypes C and B,” AIDS Research and Human Retroviruses, Vol. 21, No. 12, 2005, pp. 1016-1030. doi:10.1089/aid.2005.21.1016 [40] K. A. Buckley, P. L. Li, A. H. Khimani, R. Hof- mann-Lehmann, V. Liska, D. C. Anderson, et al., “Con- vergent Evolution of SIV Env after Independent Inocula- tion of Rhesus Macaques with Infectious Proviral DNA,” Virology, Vol. 312, No. 2, 2003, pp. 470-480. [41] S. D. Frost, Y. Liu, S. L. Pond, C. Chappey, T. Wrin, C. J. Petropoulos, et al., “Characterization of Human Immuno- deficiency Virus Type 1 (HIV-1) Envelope Variation And Neutralizing Antibody Responses during Transmission of HIV-1 Subtype B,” The Journal of Virology, Vol. 79, No. 10, 2005, pp. 6523-6527. doi:10.1128/JVI.79.10.6523-6527.2005
|