Journal of Biophysical Chemistry
Vol.2 No.4(2011), Article ID:8425,6 pages DOI:10.4236/jbpc.2011.24045
Proteomic progress in studying tuberculosis from 2010 to 2011
![]()
1Department of Pharmacogenetics, Institute of Clinical Pharmacology, Central South University, Changsha, China; hhzhou@public.cs.hn.cn
2Shanghai Public Health Clinical Center, Fudan University, Shanghai, China. *Corresponding Author: zhanglijun1221@163.com
Received 17 June 2011; revised 25 July 2011; accepted 3 August 2011.
Keywords: Proteomics; Tuberculosis; Diagnosis; Treatment
ABSTRACT
It is well accepted that rapid and early detection of Mycobacterium tuberculosis infection and understanding the mechanism of microbiologyhost interaction. Herein, we review the recently published papers related to TB proteomics from 2010 to 2011, including new technologies used in TB proteome research, diagnosis biomarkers of TB-associated diseases, disease pathogenesis and antigens for drug development. Through this review, we wish to offer some help for TB diagnosis and treatment.
1. INTRODUCTION
Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB), kills nearly two million people annually and has been a major health threat for centuries, mostly in low and middle-income countries [1-3].
However, the study for anti-TB was very slowly. One is due to no reliable biomarker for diagnosis. The gold standard for TB diagnosis is strain culture that usually costs as long as 8 weeks and the sensitivity is as low as 50% or less [4,5]. The other is no new drug. Therefore, it is very important to find new biomarkers for TB diagnosis and drug targets.
The development of proteomics has opened new ways for TB study due to its facilitation in investigating many complex issues of TB and TB-host interactions. Although proteomics has lagged behind genomics and transcriptomics due to instrumental and sensitivity problems, the application of proteomics to the study of infectious agents is beginning to emerge. Such applications include searching potential biomarkers for diagnosis [6], identifying complete virulence factor inventories [7], studying the response of both host and pathogen to the infection process (reviewed by Bhavsar AP [8] and Boshoff HI [9]), and elucidating mechanistic actions of virulence factors as they interface with host cells [10]. In this study, we reviewed proteomic progresses in TB studies from 2010 to 2011 as shown in Figure 1.
2. NEW TECHNOLOGIES DEVELOPED IN TB STUDIES
2.1. Quantitative Proteomic Studies
There are two general technologies for TB proteomic studies including: 1) two-dimensional electrophoresis (2DE) combined with mass spectrometry (2DE-MS) [11] or western blot (named 2DE-western blot(2DE-WB)) [12, 13]; 2) isotope labeling followed with multiple-dimentional liquid chromatography separation combined with mass spectrometry analysis [11]. During the last two years, a label-free proteomic method based on 1DE separation, LC-MS identification, followed by emPAI quantification was reported to identify differentially abundant proteins in closely related hypoand hyper-virulent clinical Mycobacterium tuberculosis Beijing isolates [14]. Another label-free proteomics based on SDS-PAGE separation followed by MaxQuant peak intensity calculations was developed to analyze membrane proteins of Mycobacterium tuberculosis H37Rv and H37Ra strains [7].
2.2. Proteomics for Post-Translation Modification
Studies about post-translation modifications of proteins are important for TB studies and have been reported in several studies. Pupylation is one of important post-translations which regulates protein’s degradation. Usually, pupylated proteins were affinity purified and identified by mass spectrometry [15-17]. Through expression of His7 tagged PUP and exploitation of the characteristic +243 Da mass shift attributed to tryp-
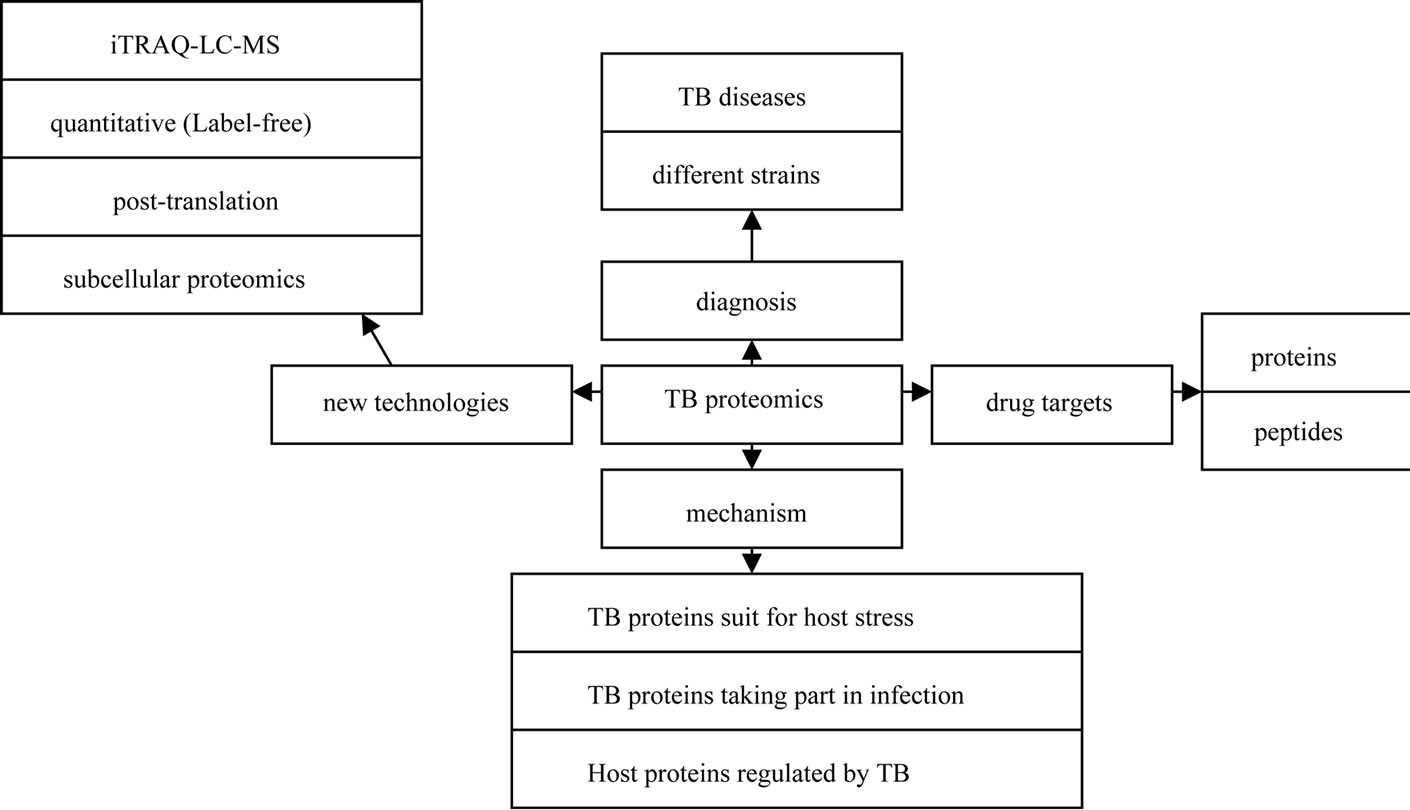
Figure 1. Illustration of proteomic progresses in TB studies from 2010 to 2011.
sinized PUPylated peptides, a global pull-down of protein targets for PUPylation in Mycobacterium smegmatis (Mycobacterium tuberculosis (Mtb)’s avirulent strain) revealed 103 candidate PUPylation targets and 52 confirmed targets[17]. Phosphorylation is another important post-translation. Prisic et al. [18] reported a phosphoproteomic study, in which they studied Ser/Thr phosphorylation in TB and found more than 500 phosphorylation events in 301 proteins.
2.3. Subcellular Proteomics
It is very difficult to study whole cell or TB proteome due to the overlap of higher abundant proteins to the lowers. In order to find proteins with low abundance or difficult to solve, subcellular proteomics was developed and used for TB study.
A proteomic study was carried in cell wall of Mycobacterium tuberculosis [19]. A total of 528 proteins were identified through a combination of detergent extraction, 2DGE, multidimensional liquid chromatography and mass spectrometry. These proteins play a role in hostpathogen interactions and define new potential drug targets via discovery of unique biosynthetic or metabolic processes. Another proteomic study was carried out in culture filtrate from M. bovis BCG Moreau, the Brazilian vaccine strain, comparing it to that of BCG Pasteur [20]. 101 differential proteins were identified, which were considered of great importance given their dominant immunogenicity and role in pathogenesis, being available for interaction with host cells since early infection. Furthermore, a proteomic strategy coupling blue native PAGE to liquid chromatography tandem mass spectrometry (LCMS/MS) was developed [21]. 40 proteins (including 12 integral membrane proteins) such as Pks7 and Pks8 were unambiguous identified. These Mycobacterial membrane and membrane-associated protein complexes play important roles in various cellular processes, and the proteinprotein interactions.
3. DISCOVERING BIOMARKERS FOR TB DIAGNOSIS
3.1. Potential Biomarkers Related to TB Infection
M. tuberculosis culturing from tissue is the gold standard for TB diagnosis, but unfortunately it takes two to six weeks for propagation. Delay in the diagnosis and treatment of TB are considered to be major factors that contribute to the high mortality of TB. Thus, from a practical as well as a research standpoint, development of new markers for TB is desired. Proteomics, due to its high-throughout and sensitivity, has been widely used to find potential biomarkers for TB diagnosis [6,11,22,23]. Liu Q [6] used SELDI-TOF to analyze a total of 155 sera samples from TB patients and its controls. They found that mass to charge ratios (m/z) of 4821.45, 3443.22, and 9284.93, etc. determined in the training set could detect 93.55%, 95.46%, and 88.09% accuracy for classifying smear-positive pulmonary tuberculosis (SPPTB), smearnegative pulmonary tuberculosis (SNPTB) patients and non-tuberculosis (non-TB) controls, respectively. Kashyap RS [22] found that the 65-kDa HSP, 71-kDa HSP, 14-kDa HSP and Ag 85 complex proteins were overexpressed in TB ascites compared with non-TB ascites patients through proteomic study. Takahiro Tanaka [24] found that RBP4 and fetuin-A were significantly lower expressed in samples from patients with active TB than in their controls (P < 0.0001) through analyzing the whole blood supernatant.
3.2. Potential Biomarkers to Distinguish Different Strains
Except diagnosing TB infection, the ability to readily and accurately distinguish among strains is important because different strains have different virulence and drugresistances characters that necessitate different treatments. A label-free quantitative proteomic approach [23] showed that 48 proteins were over-represented in the hypo-virulent isolate, while 53 were overrepresented in the hyper-virulent. These proteins such as ESAT-6, Esxlike proteins and fbpD (MPT51) reduced in the hypervirulent strain might be used for distinguishing hypo and hyper-virulent TB. Similarly, a label-free quantitative proteomic approach was performed in the virulent H37- Rv strain and its attenuated counterpart H37Ra. The results showed that 29 membrane-associated proteins with a 5 or more fold difference in their relative abundance in one strain compared to the other. Of which, 19 membraneand lipo-proteins such as three ABC transporter proteins (Rv0933, Rv1273c and Rv1819c) had higher abundance in H37Rv, while another 10 proteins had a higher abundance in H37Ra [7]. Furthermore, Mehaffy et al. [11] studied the proteomes of secreted and cytosolic proteins of genetically closely related strains of M. tuberculosis through 2DE and iTRAQ-LC-MS methods, and found that some enzymes such as GltA2, SucC, Gnd1 and Eno were expressed differently in different strains.
4. MECHANISM OF TB INFECTION
Although TB has been found for a long time, the knowledge about TB infection is still limited. Proteomics as a new technology has shown great advance in the study of TB infection.
4.1. TB Proteins Suit for Host Stress
During the TB infection, the tubercle bacilli are likely to be exposed to stress that can result in the formation of aberrant proteins with altered structures. Bacteria have evolved accurate regulatory systems to control the expression and function of potentially destructive proteases and chaperones. For example, the Clp Gene Regulator(ClgR) of M.tuberculosis activates the transcription of at least ten genes including four of which encode protease systems (ClpP1/C, ClpP2/C, PrtB and an HtrA-like protease-Rv1043c) and three of which encode chaperones (Acr2, ClpB and Rv3269) [25]. HtrA-like serine protease encoded by pepD, are responsible for degrading or refolding protein substrates following exposure to stress. A proteomic study further showed that the HtrA-Like Serine Protease PepD interacts with and modulates the Mycobacterium tuberculosis 35-kDa antigen outer envelope protein [26].
4.2. TB Proteins Taking Part in Infection
Considerable effort has been directed toward controlling tuberculosis through understanding host-pathogen interactions leading to biomarker expression. During the early and chronic stages of disease, it is proteins from TB that regulates host cells. Through a guinea pig model of tuberculosis, 500 unique proteins were identified in the infected lung. Of which cell wall and cell wall processes, and intermediary metabolism and respiration were the two major functional classes, including Rv2209, Rv2315c, etc [27]. Kunnath VS [12] also found that sera from worldwide TB suspects recognized approximately 10% of the bacterial proteome and the M. tuberculosis immunoproteome were rich in membrane-associated and extracellular proteins through carrying a systems immunology approach.
4.3. Host Proteins Regulated by TB
TB can regulate host proteome to finish its infection. Proteins involved in membrane trafficking and signal transduction such as Ras GTPase-activating-like protein IQGAP1 were detected to be regulated through a proteomic study [28]. In this study, the phagosomes from BCGinfected human macrophages were purified, and 447 human host proteins were identified. Furthermore, Mycobacteria release active membrane vesicles (MVs) to deliver ligands that can be recognized by host cells. Extensive proteomic analysis revealed that only MVs from the virulent strains contained TLR2 lipoprotein agonists. The interaction of MVs with macrophages isolated from mice stimulated the release of cytokines and chemokines in a TLR2-dependent fashion [29].
Except the whole TB can regulate host proteome, some function proteins such as lipoarabinomannan (LAM) that were found to change host protein’s expression through proteomic studies [10]. LAM (the mycobacterial cell wall component) has been described as one of the key virulence factors of Mycobacterium tuberculosis. Through treating the macrophages with LAMs purified from the two mycobacterial species, protein trafficking pathways that contribute to the arrest of phagosome maturation were found to be regulated [10].
5. PROTEOMICS IN DRUG DEVELOPMENT
5.1. Proteins for Drug Development
The inadequate protection afforded by bacillus Calmette-Guérin (BCG) vaccination provides continued impetus for the discovery of immunodominant Mycobacterium tuberculosis (MTB) antigens to develop improved vaccines. The 6-kDa early secretory antigenic target of Mycobacterium tuberculosis (ESAT-6) [30] and the 10- kDa culture filtrate antigen (CFP-10) [31] are the most immunodominant and highly M. tuberculosis (MTB)-specific antigens. Ag85B-ESAT-6 adjuvanted with IC31 promotes strong and long-lived Mycobacterium tuberculosis specific T cell responses in naive human volunteers [30]. Rv3615c [Esx-1 substrate protein C (EspC)], a protein with similar size and sequence to CFP-10 and ESAT- 6, was found to be a highly immunodominant RD1 (Region of Difference 1)-dependent secreted antigen specific for Mycobacterium tuberculosis infection through a quantitative proteomics and metabolically labeled mutant and genetically complemented MTB strains [32]. Giri et al. [33] used LC-MS/MS to identify 41 mycobacterial proteins present in exosomes released from M. tuberculosis-infected J774 cells. Another quantitative proteomic analysis showed a higher expression of immunogenic proteins such as Rv1860 (BCG1896, Apa), Rv1926c (BCG- 1965c, Mpb63) and Rv1886c (BCG1923c, Ag85B) in BCG Moreau when compared to BCG Pasteur from their culture filtrate [20]. These antigens might offer new targets for vaccine development. Nearly, an immunoproteome microarray study of M. tuberculosis-proteins with sera from patients was developed [34], in which, 3 novel antigens, namely, Rv1987, Rv3807c, and Rv3887c, provided better sensitivity and accuracy for TB detection.
Sarah L. Kinnings built a TB-drugome(http://funsite.- sdsc.edu/drugome/TB) database. In this TB-drugome, 123 of the 274 drugs are connected to 447 of the 1730 proteins. These proteins are the potential targets for developing some safe and efficient anti-tubercular drugs [35].
5.2. Peptides for Drug Development
It is proposed that low-similarity peptides be a potential epitope against M. tuberculosis. The TB sequence were examined for similarity score to the proteins of the host in which the epitopes were defined [36]. Additionally, a peptide microarray consisting of 7466 unique peptides derived from 61 M. tuberculosis proteins for potential MHC class II-presented epitopes was constructed. Two hundred and twenty-two peptides that formed MHC class II-peptide complexes had previously been described as exclusively recognized by IgG in sera from patients with active pulmonary tuberculosis [37].
6. CONCLUSIONS
This review summarized the new proteomic technologies used in anti-TB studies and provided a better understanding for possible new biomarkers and infection mechanism as well as drug development in TB study through proteomic technologies. Although the proteins provided by proteomic studies might not be used for biomarkers or drug targets at once, they will be helpful for anti-TB research and must promote TB treatment development through further studies. In all, proteomics may open up new horizons for understanding the pathogenesis of tuberculosis and developing drugs.
7. ACKNOWLEDGEMENTS
This work was supported by “China Postdoctoral Science Foundation (20100471238)”, the National Basic Research Program of China (also called 973 Program) (2011CB910700) and the Postdoctoral Science Foundation of Central South University.
REFERENCES
- Prasad, R. (2010) Multidrug and extensively drug-resistant TB (M/XDR-TB): Problems and solutions. Indian Journal of Tuberculosis, 57, 180-191.
- Ahmad, S. (2010) New approaches in the diagnosis and treatment of latent tuberculosis infection. Respiratory Research, 11, 169. doi:10.1186/1465-9921-11-169
- (2010) WHO global tuberculosis control report 2010. Summary. Central European Journal of Public Health, 18, 237.
- Sendagire, I., Schim Van der Loeff, M., Mubiru, M., Konde-Lule, J. and Cobelens, F. (2010) Long delays and missed opportunities in diagnosing smear-positive pulmonary tuberculosis in Kampala, Uganda: A cross-sectional study. PLoS One, 5, e14459. doi:10.1371/journal.pone.0014459
- Bailey, S.L., Roper, M.H., Huayta, M., Trejos, N., Lopez Alarcon, V. and Moore, D.A. (2010) Missed opportunities for tuberculosis diagnosis. The International Journal of Tuberculosis and Lung Disease, 15, 205-210.
- Liu, Q., Chen, X., Hu, C., Zhang, R., Yue, J., Wu, G., et al. (2010) Serum protein profiling of smear-positive and smear-negative pulmonary tuberculosis using SELDITOF mass spectrometry. Lung, 188, 15-23. doi:10.1007/s00408-009-9199-6
- Malen, H., De Souza, G.A., Pathak, S., Softeland, T. and Wiker, H.G. (2011) Comparison of membrane proteins of Mycobacterium tuberculosis H37Rv and H37Ra strains. BMC Microbiology, 11, 18. doi:10.1186/1471-2180-11-18
- Bhavsar, A.P., Auweter, S.D. and Finlay, B.B. (2010) Proteomics as a probe of microbial pathogenesis and its molecular boundaries. Future Microbiology, 5, 253-265. doi:10.2217/fmb.09.114
- Boshoff, H.I. and Lun, D.S. (2010) Systems biology approaches to understanding mycobacterial survival mechanisms. Drug Discovery Today: Disease Mechanisms, 7, 75-82. doi:10.1016/j.ddmec.2010.09.008
- Shui, W., Petzold, C.J., Redding, A., Liu, J., Pitcher, A., Sheu, L., et al. (2011) Organelle membrane proteomics reveals differential influence of mycobacterial lipoglycans on macrophage phagosome maturation and autophagosome accumulation. Journal of Proteome Research, 10, 339-348. doi:10.1021/pr100688h
- Mehaffy, C., Hess, A., Prenni, J.E., Mathema, B., Kreiswirth, B. and Dobos, K.M. (2010) Descriptive proteomic analysis shows protein variability between closely related clinical isolates of Mycobacterium tuberculosis. Proteomics, 10, 1966-1984. doi:10.1002/pmic.200900836
- Kunnath-Velayudhan, S., Salamon, H., Wang, H.Y., Davidow, A.L., Molina, D.M., Huynh, V.T., et al. (2010) Dynamic antibody responses to the Mycobacterium tuberculosis proteome. Proceedings of the National Academy of Sciences of the United States of America, 107, 14703- 14708. doi:10.1073/pnas.1009080107
- Deenadayalan, A., Heaslip, D., Rajendiran, A.A., Velayudham, B.V., Frederick, S., Yang, H.L., et al. (2010) Immunoproteomic identification of human T cell antigens of Mycobacterium tuberculosis that differentiate healthy contacts from tuberculosis patients. Molecular & Cellular Proteomics, 9, 538-549. doi:10.1074/mcp.M900299-MCP200
- de Souza, G.A., Fortuin, S., Aguilar, D., Pando, R.H., McEvoy, C.R., van Helden, P.D., et al. (2010) Using a label-free proteomics method to identify differentially abundant proteins in closely related hypoand hypervirulent clinical Mycobacterium tuberculosis Beijing isolates. Molecular & Cellular Proteomics, 9, 2414-2423. doi:10.1074/mcp.M900422-MCP200
- Festa, R.A., McAllister, F., Pearce, M.J., Mintseris, J., Burns, K.E., Gygi, S.P. and Darwin, K.H. (2010) Prokaryotic ubiquitin-like protein (Pup) proteome of Mycobacterium tuberculosis [corrected]. PLoS One, 5, 8589. doi:10.1371/journal.pone.0008589
- Poulsen, C., Akhter, Y., Jeon, A.H., Schmitt-Ulms, G., Meyer, H.E., Stefanski, A., et al. (2010) Proteome-wide identification of mycobacterial pupylation targets. Molecular Systems Biology, 6, 386. doi:10.1038/msb.2010.39
- Watrous, J., Burns, K., Liu, W.T., Patel, A., Hook, V., Bafna, V., et al. (2010) Expansion of the mycobacterial “PUPylome”. Molecular Biosystems, 6, 376-385. doi:10.1039/b916104j
- Prisic, S., Dankwa, S., Schwartz, D., Chou, M.F., Locasale, J.W., Kang, C.M., et al. (2010) Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proceedings of the National Academy of Sciences of the United States of America, 107, 7521-7526. doi:10.1073/pnas.0913482107
- Wolfe, L.M., Mahaffey, S.B., Kruh, N.A. and Dobos K.M. (2010) Proteomic definition of the cell wall of Mycobacterium tuberculosis. Journal of Proteome Research, 9, 5816-5826. doi:10.1021/pr1005873
- Berredo-Pinho, M., Kalume, D.E., Correa, P.R., Gomes, L.H., Pereira, M.P., Silva, R.F., et al. ( 2011) Proteomic profile of culture filtrate from the Brazilian vaccine strain Mycobacterium bovis BCG Moreau compared to M. bovis BCG Pasteur. BMC Microbiology, 11, 80. doi:10.1186/1471-2180-11-80
- Zheng, J., Wei, C., Zhao, L., Liu, L., Leng, W., Li, W. and Jin, Q. (2011) Combining blue native polyacrylamide gel electrophoresis with liquid chromatography tandem mass spectrometry as an effective strategy for analyzing potential membrane protein complexes of Mycobacterium bovis bacillus Calmette-Guerin. BMC Genomics, 12, 40. doi:10.1186/1471-2164-12-40
- Kashyap, R.S., Saha, S.M., Nagdev, K.J., Kelkar, S.S., Purohit, H.J., Taori, G.M. and Daginawala, H.F. (2010) Diagnostic markers for tuberculosis ascites: A preliminary study. Biomark Insights, 5, 87-94.
- Desouza, G.A., Fortuin, S., Aguilar, D., Pando, R.H., Mc Evoy, C.R., van Helden, P.D., et al. (2010) Using a label-free proteomic method to identify differentially abundant proteins in closely related hypoand hyper-virulent clinical Mycobacterium tuberculosis Beijing isolates. Molecular & Cellular Proteomics, 9, 2414-2423. doi:10.1074/mcp.M900422-MCP200
- Tanaka, T., Sakurada, S., Kano, K., Takahashi, E., Yasuda, K., Hirano, H., et al. (2011) Identification of tuberculosis-associated proteins in whole blood supernatant. BMC Infectious Diseases, 11, 71. doi:10.1186/1471-2334-11-71
- Estorninho, M., Smith, H., Thole, J., Harders-Westerveen, J., Kierzek, A., Butler, R.E., et al. (2010) ClgR regulation of chaperone and protease systems is essential for Mycobacterium tuberculosis parasitism of the macrophage. Microbiology, 156, 3445-3455. doi:10.1099/mic.0.042275-0
- White, M.J., Savaryn, J.P., Bretl, D.J., He, H., Penoske, R.M., Terhune, S.S. and Zahrt, T.C. (2011) The HtrA-like serine protease PepD interacts with and modulates the mycobacterium tuberculosis 35-kDa antigen outer envelope protein. PLoS One, 6, 18175. doi:10.1371/journal.pone.0018175
- Kruh, N.A., Troudt, J., Izzo, A., Prenni, J. and Dobos, K.M. (2010) Portrait of a pathogen: The Mycobacterium tuberculosis proteome in vivo. PLoS One, 5, 13938. doi:10.1371/journal.pone.0013938
- Lee, B.Y., Jethwaney, D., Schilling, B., Clemens, D.L., Gibson, B.W. and Horwitz, M.A. (2010) The mycobacterium bovis bacille calmette-guerin phagosome proteome. Molecular & Cellular Proteomics, 9, 32-53. doi:10.1074/mcp.M900396-MCP200
- Prados-Rosales, R., Baena, A., Martinez, L.R., LuqueGarcia, J., Kalscheuer, R., Veeraraghavan, U., et al. (2011) Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. The Journal of Clinical Investigation, Epub ahead of print. doi:10.1172/JCI44261
- van Dissel, J.T., Arend, S.M., Prins, C., Bang, P., Tingskov, P.N., Lingnau, K., et al. (2010) Ag85B-ESAT-6 adjuvanted with IC31 promotes strong and long-lived Mycobacterium tuberculosis specific T cell responses in naive human volunteers. Vaccine, 28, 3571-3581. doi:10.1016/j.vaccine.2010.02.094
- Dannenberg, A.M. Jr. (2010) Perspectives on clinical and preclinical testing of new tuberculosis vaccines. Clinical Microbiology Reviews, 23, 781-794. doi:10.1128/CMR.00005-10
- Millington, K.A., Fortune, S.M., Low, J., Garces, A., Hingley-Wilson, S.M., Wickremasinghe, M., et al. (2010) Rv3615c is a highly immunodominant RD1 (Region of Difference 1)-dependent secreted antigen specific for Mycobacterium tuberculosis infection. Proceedings of the National Academy of Sciences of the United States of America, 108, 5730-5735. doi:10.1073/pnas.1015153108
- Giri, P.K., Kruh, N.A., Dobos, K.M. and Schorey, J.S. (2010) Proteomic analysis identifies highly antigenic proteins in exosomes from M. tuberculosis-infected and culture filtrate protein-treated macrophages. Proteomics, 10, 3190-3202. doi:10.1002/pmic.200900840
- Li, Y., Zeng, J., Shi, J., Wang, M., Rao, M., Xue, C., et al. (2010) A proteome-scale identification of novel antigenic proteins in Mycobacterium tuberculosis toward diagnostic and vaccine development. Journal of Proteome Research, 9, 4812-4822. doi:10.1021/pr1005108
- Kinnings, S.L., Xie, L., Fung, K.H., Jackson, R.M., Xie, L. and Bourne, P.E. (2010) The Mycobacterium tuberculosis drugome and its polypharmacological implications. PLoS Computational Biology, 6, 1000976. doi:10.1371/journal.pcbi.1000976
- Lucchese, G., Stufano, A. and Kanduc, D. (2010) Proposing low-similarity peptide vaccines against Mycobacterium tuberculosis. Journal of Biomedicine and Biotechnology, 832341.
- Gaseitsiwe, S., Valentini, D., Mahdavifar, S., Reilly, M., Ehrnst, A. and Maeurer, M. (2010) Peptide microarray-based identification of Mycobacterium tuberculosis epitope binding to HLA-DRB1*0101, DRB1*1501, and DRB1* 0401. Clinical and Vaccine Immunology, 17, 168- 175. doi:10.1128/CVI.00208-09

