Paper Menu >>
Journal Menu >>
 Engineering, 2012, 5, 167-171 doi:10.4236/eng.2012.410B044 Published Online October 2012 (http://www.SciRP.org/journal/eng) Copyright © 2012 SciRes. ENG Cloni ng, Expression, and Immunogenicity Analysis of the CpsE Protein from Group B Streptococcus Isolated from Tilapia (Oreochromis Niloticus) Xi Fu1, Kaiyu Wang1, Jinlu Huang1, Jun Wang1, H ai Lian1, Yang He1, Lanmin Li1, Kaiyu Wang2 1Fish Disease Research Cent er, C o l lege of Vet er in ary Medicine o f Sich uan Agri cu ltu r al University, Y a’a n,China 2Key Laboratory of Animal Di sease and Human Health of Sichuan P rovince, College of Veterinary Medicine of Sichuan Agricultural University, Ya’an, China Email: kywangsicau@126.com Received 2012 ABSTRACT Group B St reptoco ccus (GBS) is a major cause o f seriou s bacterial in fection in nu merous ani mal species. The prod uction o f capsular polysaccharide(CPs) is vital to GBS to evade host immunity. One of the genes that required for production of CPs, cpsE, has been determined to be well conserved in capsule gene cluster (cps).This study cloned the cpsE gene from Tilapia of GBS clinical isolate (serotype Ia) and expressed this gene with aid of pET-32a(+) in Escheri chia co li BL21(DE3) competent cells to obtain high levels of the recombinant protein for further study about CpsE in fish and examination of its immunogenicity. The optimization of induction conditions (IPTG concentration, temperature and time) in E.coli was accomplished and let us to perform the recombinant protein induction at 37℃ for 3h,with 0.2mM IPTG in Luria Bertani (LB) medium. At the optimal conditions, recombinant protein was ex- pressed in an insoluble form (inclusion bodies) and accounted for approximately 23% of the total protein. Purification by affinity chromatography yielded about 480mg fusion protein per liter culture. Keywords: Recombinant CpsE Expression ; Purification; Poly clonal Antibody 1. Introduction Group B Streptococcus (GBS) is the foremost cause of life-threatening bacterial infection in human and other animals [1-4] . GBS also causes septicaemia and meningitis of obvious morbidity and mortality in fish such as tilapia (Oreochromis niloticus), cultured seabream, and wild mullet [5-7]. It is com- monly bel ieved that t he threat of GBS disease is a considerable health burden for society. For GBS, CPs is a major virulence factor as well as a target for protective immunity [8].To dat e, ten serotypes (serotypes Ia, Ib, and II through IX) of GBS have been identified based on their unique CPS antigens[10-12]. Among these serot yp es , I a、 III、V and VI I I are prevalent among human [11,13,14], the most currently isolated from fish are Ia , Ib and III [15,1 6]. There are five genes (cpsA to –E) that are well conserved among sever al po lysacchari de-producing streptococci including S. thermophilus, S. pneumoniae, S. suis and among the GBS capsule serotypes[17-19]. According to Cieslewicz et al[20], inactivation of cpsIaE resulted in an acapsular phenotype, con- sistent with previous work that identified CpsE as the glycosyl- transferase [18,22]. Impact of GBS infection on fish was sporadically reported throughout the world [5,6,23-25] . Here we report molecular cloning、prokaryotic expression and purification of the recom- binant protein. Additionally, we studied the optimal induction conditions for recombinant protein including concentration of IPTG (Isopropyl b-D-1-thiogalactopyranoside), induction tem- peratures and induction time. 2. Materials and Methods 2.1. Clinic al Isolate, Plasmids and Cultur e Con ditions GBS strains (n=10 ) were isolated fro m liver、brain and kidney from 100 infected tilapia (Wen-chang City, Hainan Prov- ince ,China). One virulent strain was selected through regres- sion test over healthy tilapia. The strain was identified to ex- press the type Ia capsule by multiplex PCR assay [26]. All E.coli clones were routinely grown in LB broth containing 100μg/ml of ampicillin at 37℃. 2.2. Translated Protein Analysis The Pro tean program of so ftware DNAStar was used t o predict the secondary structure and the surface probability as well as antigenic index of translated protein. SOSUI Server (h ttp ://bp. nuap.nagoya-u.ac.jp/sosui/sosui_submit.html) and SignalP 4.0 Server (http://www.cbs.dtu.dk/services/SignalP/) were used to analyze th e presence of tr ansmembran e region s and signal pep- tide, respectively. The hydrophilicity character of the protein sequence was predicted by software Bioedit7.0.Chance of in- solubility of target protein over-expressed in E.coli was pre- dicted using online program (http://www.biotech.ou.edu/). 2.3. Primer Design and PC R Amplificatio n of the cpsE Gene The full-length of cpsE gene in this study (GenBank accession no.HQ888682) consists of 450 base pairs. Primers with restric-  X. FU ET AL. Copyright © 2012 SciRes. ENG 168 tion sites Bam HI and Hind III (underlined), respectively. Pri- mers: upstream 5'- CGGGATCCATGAAAATTTGTCTGGTTGG-3’ downstream 5'- CCAAGCTT Bacterial isolates from infected tilapia were initially identified as GBS by conventional phenotypic characteristics (Gram- positive cocci, β-haemolytic, catalase negative, API 20 STREP system, Slidex Strepto-kit). Slidex Strepto-kit was positive for group B and API 20 STREP gave profile number 3663414, which co r r es ponded to excellent matches to GBS. 3.2. Bioinformatics Prediction The analysis results obtained by the software DNAStar indi- cated that there were nine distinct antigenic domains and six surface probability domains. This program also showed a mea- surable amount of beta strands with few turn regions and ran- dom coil. Prediction of SignalP 3.0 Server confirmed the ab- sence of signal peptide in CpsE sequence. Program SOSUI Server indicated that amino acids from 80-102aa in the cpsE gene may be part of a transmembrane region in the CpsE pro- tein sequence .Results indicate that this region (80-102aa) is likely anchored to the bacterial wall. And possibility of insolu- bility when over-expressed in E.coli was 64.1%. 3.3. Plasmids Con struction Coning plasmid pMD19-t/cpsE was confirmed by double re- striction enzymes digestion (Bam HI and Hind III) (Figure 1) and sequencing analysis. Correct fragment was fused with pET-32a (+) to form the expression plasmid pET-32a (+)/cpsE. The initial transformation was performed with competent DH5αcells for screening. The positive clones were identified by PCR amplification and double restriction enzymes digestion (Figure 2). TTAAAAAATTCCTCCTAAATT-3’). Amplification was done at 94℃ for 5 min and then at 94℃ for 1 min, 55℃for 1 min, and 72℃ for 40 s for a total of 30 cycles, followed by 72℃ for 10 m in. 2.4. Construction of Cloning Plasmid and Expre ssion Plasmids The target fragment was ligated into cloning vector pMD19-T to construct recombinant cloning plasmid pMD19-T/cpsE. Competent E.coli strain DH5α cells were used for host cloning construction. The pET-32a (+) vector and the recombinant cloning plas- mids were both digested with Bam HI and Hind III and were both spliced with T4 ligase (Takara, Japan) to construct the recombinant prokaryotic expression plasmid pET-32a(+)/ cpsE. The ligated product was transformed into competent DH5a cells . Selection of positive clones was done by PCR amplification, double enzymes digestion and sequence analysis. The positive clones were transformed into expression hosts of BL21 (DE3) cells. 2.5. Expression and O ptimization o f Expression Conditions Transformants were inoculated into 5 ml of culture medium in test tubes and grown overnight at 37℃with constant agitation (220 rpm) until they reached the exponential phase (approx- imately OD600≈ 0.5–0.6 measured by absorbance at 600nm) and were subsequently induced with IPTG. Preliminary tests were performed with 1mM IPTG induction at 37 °C for 4 h and for the optimization of mature CpsE expression. Optimization of the expression conditions were performed at different tempera- tures (30℃, 34℃, and 37℃), for different induction periods (1h, 2h, 3h, 4h, 5h, 6h, 7 h and overnight) and with different IPTG concentrations (0.2mM, 0.4mM. 0.6mM, 0.8mM, 1.0mM, 1.2mM, 1.5mM and 2.0mM). 2.6. Purification of the His6-tagged Recombinant Protein The His6-tagged recombinant protein was subjected to Bio- RadTM metal affinity chromatography for purification. Under denatured state, CpsE-containing fractions were pooled and dialyzed against PBS buffer (10mM Na2 HPO 4 1.8mM KH 2 PO4, pH 7.4, 140mM NaCl, and 2.7mM KCl) with step-wise lower con centration s of urea (6M, 4M, 2M and 0M ) for a total time of 96h. The purified recombinant protein was stored at 4℃ for use within 1 week or at –70℃ for longer time. 3. Results 3.1. GBS Strain Isolate Confirming Figure1. Characterization of the recombinant plasmid pMD19- T/cpsE by restriction digestion and PCR-based amplification.M1: DNA marker of 7000bp; lane 1:pMD19-T/cpsE digested with Bam HI; lane 2:pMD19-T/cpsE dig e s t e d wi t h Bam H I an d Hind III; la ne 3:PCR amplification product of cpsE (ORF450bp); M2:DNA m arker of 200 0b p Fi gur e 2. Characterization of the recombinant plasmid pET-32a(+)/cp sE by restriction digestion and PCR-based amplifi- cation. M1:DNA marker of 200bp;lane 1:PCR amplification prod- uct of cpsE ; lane 2:pET-32a(+)/cpsE digested with Bam HI and Hind III; lane 3: pET-32a(+)/c p sE digested with Bam HI; lane 4: pET-32a(+)digested with Bam HI and Hind III; lane 5: pET-32a(+) digested with Bam HI. 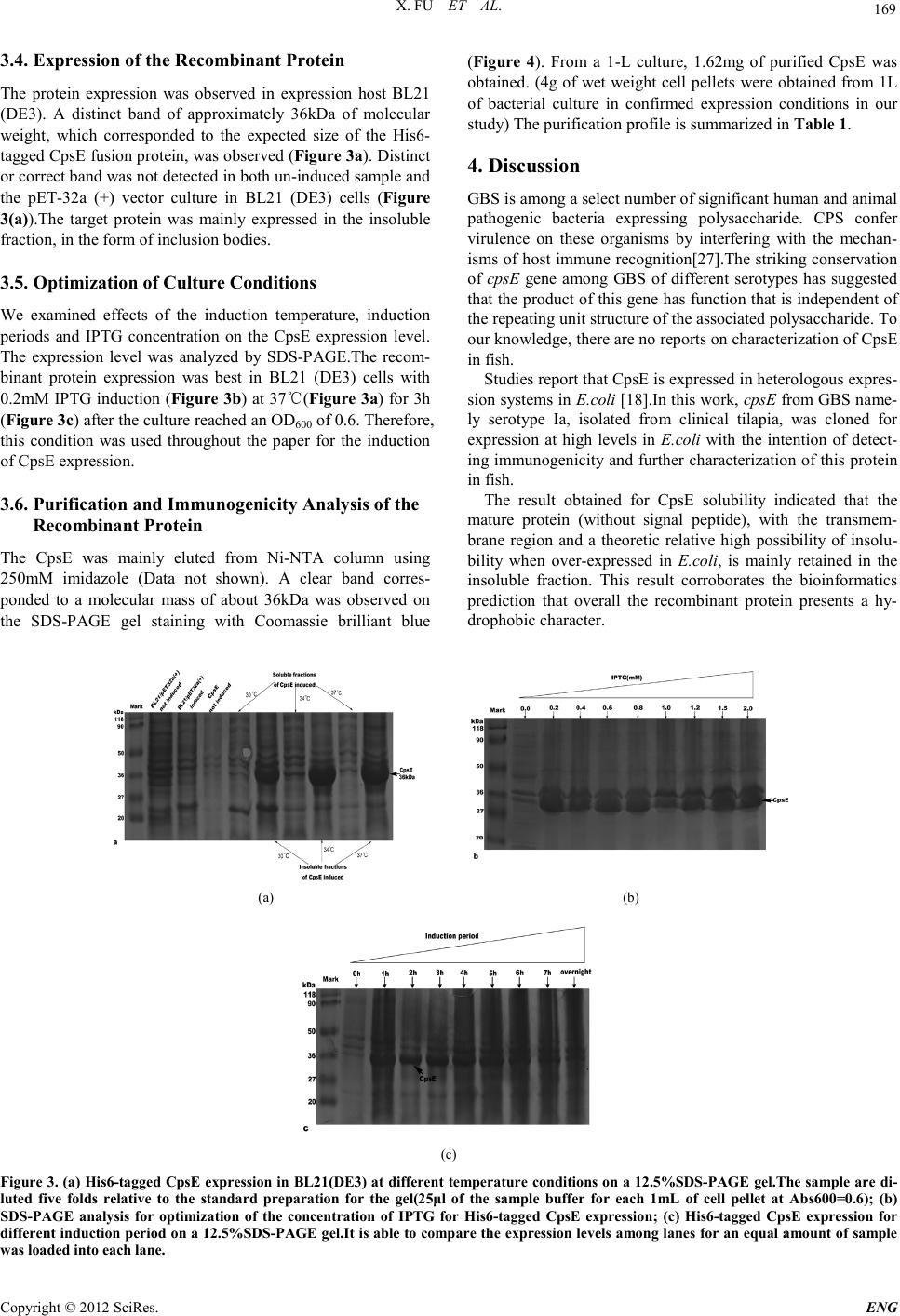 X. FU ET AL. Copyright © 2012 SciRes. ENG 169 3.4. Expression of the Reco mbinant Protein The protein expression was observed in expression host BL21 (DE3). A distinct band of approximately 36kDa of molecular wei g h t , which corresponded to the expected size of the His6- tagged CpsE fusion protein, was observed (Figure 3 a). Distinct or correct band was not detected in both un-induced sample and the pET-32a (+) vector culture in BL21 (DE3) cells (Figure 3(a)).The target protein was mainly expressed in the insoluble fraction, in the form of inclusion bodies. 3.5. Optimization of Cul ture Conditions We examined effects of the induction temperature, induction periods and IPTG concentration on the CpsE expression level. The expression level was analyzed by SDS-PAGE.The recom- binant protein expression was best in BL21 (DE3) cells with 0.2mM IPTG induction (Figure 3b) at 3 7℃(Figure 3a) for 3h (Figure 3c) after the cult ure r eached an OD600 of 0.6. Therefore, this condition was used throughout the paper for the induction of CpsE expression. 3.6. Purification and Immunogenicity Analysis of the Reco mbinant Prote in The CpsE was mainly eluted from Ni-NTA column using 250mM imidazole (Data not shown). A clear band corres- ponded to a molecular mass of about 36kDa was observed on the SDS-PAGE gel staining with Coomassie brilliant blue (Figure 4). From a 1-L culture, 1.62mg of purified CpsE was obtained. (4g of wet weight cell pellets were obtained from 1L of bacterial culture in confirmed expression conditions in our study) The purification profile is summarized in Table 1. 4. Discussion GBS is among a select number of significant human and animal pathogenic bacteria expressing polysaccharide. CPS confer virulence on these organisms by interfering with the mechan- isms of host immune recognition[27] .The striking conservation of cpsE gene among GBS of different serotypes has suggested that the product of this gene has function that is independent of the repeating unit structure of the associated polysaccharide. To our knowledge, there are no reports on characterization of CpsE in fish. Studies report that CpsE is expressed in heterologous expres- sion systems in E.coli [18].I n this work, cpsE from GBS name- ly serotype Ia, isolated from clinical tilapia, was cloned for expression at high levels in E.coli with the intention of detect- ing immunogenicity and further characterization of this protein in fish. The result obtained for CpsE solubility indicated that the mature protein (without signal peptide), with the transmem- brane region and a theoretic relative high possibility of insolu- bility when over-expressed in E .coli, is mainly retained in the insoluble fraction. This result corroborates the bioinformatics prediction that overall the recombinant protein presents a hy- drophobic ch aracter. (a) (b) (c) Figure 3. (a) His 6 -tagged CpsE expression in BL21(DE3) at differe nt temperature conditions on a 12.5%SDS-PAGE gel.The sample are di- luted five folds relative to the standard preparation for the gel(25μl of the sample buffer for each 1mL of cell pellet at Abs600=0.6); (b) SDS-PAGE analysis for optimization of the concentration of IPTG for His6-tagged CpsE expression; (c) His6 -tagged CpsE expression for differe nt induction period o n a 12.5%SDS-PAGE gel.It is able to compare the expression levels among lanes for an equal amount of sample was lo a d ed in to ea ch lane.  X. FU ET AL. Copyright © 2012 SciRes. ENG 170 Fi gur e 4. Purified CpsE on a 12.5%SDS-PAGE gel eluted from Ni-NTA column using 250mM imidazole. Expression of all lanes ar e a cco mplished at optimal co nditions (at 37℃ for 1h with 0.2mM IPTG induction). Table 1. Purification of cpse protein from 1l culture. Step Total proteina (mg) CpsE (mg) Protein concentration μg/mlb Purity (%)c Cru de ext ractd 67.5 15.48 1,214 23 Immobilized metal affinity chromatography 1.90 1.62 480 85 aTotal protein was isolated from 1liter of culture medium in optimal expression condition; b Protein concentration was estimated by the Bradford method using BSA as standard; c The purity was determined as the amount of the stain asso- ciated with the CpsE band as a fraction of the stain associated with all the bands on the SDS-PAGE[28]; d The pellets containing the insoluble fraction (crude extract) obta ine d from 1liter of culture. To date, there are no reports in literature that ap proached the expression optimization allowing the development of a process to produce recombinant CpsE in E.coli. Our work is able to present conditions that are optimal for production of recombi- nant protein. Distribution of B cell epitopes is largely related to the secondary structures of the protein because regions of turn and coil can easily change in shape that benefits combination of protein with antibodies. Hydrophilic regions of the protein would serve as likely B cell epitopes. Prediction results of hy- drophilicity character and transmembrane region indicate that there may have certain B cell epitopes in the CpsE protein. Comprehensive analysis of turn regions, coil regions, surface probability regions and hydrophilic regions presented here suggest that the most possible sites for B cell epitopes are Phe52-Asn57, Gly7-Gly11, Ser83-Gly84 and Asp112-Pro 114. This speculate is able to locate B cell epitopes, which could help to find sp ecific antigen and furth er benefit development of subunit vaccin e. 5. Acknowledgements This work was supported by grant from Program for Chang- jiang Scholars and Innovative Research Teamin in University (PCSIRT) (Grant no. IRT0848 ); Science and Technology project of Sichuan Province (No.08ZA082). Kaiyu Wang is the corresponding author. Fish Disease Research Center & Key Laboratory of Animal Disease and Human Health of Sichuan Province, College of Veterinary Medicine of Sichuan Agri- cultural University, 46# Xinkang Road, Yucheng District, Yaan 62501 4, Si c h ua n Pr ovince of Chi na . REFERENCES [1] S.J.Schrag, D.Phil, E.R.Zell, M.Stat, R.Lynfield, A.Roome, K.E.Arnold, A.S.Craig, L.H.Harrison, A.Reingold, K.Stefonek, G.Smith, M.Gamble, A.Schuchat, “A population-based compar- ison of strategies to prevent early-onset group B streptococcal disease in neonates,” N. Engl. J. Med. New York, vol.347, pp.233-239, July 2002. [2] M.S.Edwards, C.J.Baker, “Group B streptococcal infections in elderly adults,” Clin. Infect. Dis. New York, vol.41 pp.839-847, May 2005. [3] T.H.Skoff, M.M.Farley, S.Petit, A.S.Craig, W.Schaffner, K.Gershman, L.H.Harrison, R.Lynfield, J.M.Boetani, S.Zansky, B.A.Albanese, K.Stefonek, E.R.Zell, D.Jackson, T.Thompson, S.J.Schrag, “Increas ing bu rden of in vasive group B streptoc occal dis ease in nonpregna nt adults 199 0-2007,”Clin. Infect. Dis. New York,vol.49 pp.85-92, February 2009. [4] G.P.Keefe, “Streptococcus agalactiae mastitis: a review,” Can.Vet.J. Canada,vol.38, pp.429-437,July 1997. [5] J.J.Evans, P.H.Klesius, P.M.Gilbert, C.A.Shoemaker, M.A.A.Sarawi, J.Landsberg, R.Duremdez, “Characterization of β-haemolytic group B streptococcus agalactiae in cultured sea- bream, Sparus auratus L., and wild mullet, Liza klunzingeri (Day), in Kuwait,” J.Fish. Dis.Wiley, New York, vol.25, pp.505-513, September 2 002. [6] R.Salvador, E.E.Muller, J.C.Freitas, J.H.Leonhadt, “Isolation and characterization of Streptococcus spp. group B in Nile tilapia (Oreochromis niloticus) reared in hapas nets and earth nurseries in the northern region of Parana State,Brazil,” Ciência Rural Santa Mar ia, vol.35,pp.1374-1378, November 2005. [7] A.Eldar, Y.Bejerano, H.Bercovier, “Streptococcus shiloi and Streptococcus difficile: two new streptococcal species causing a meningoencephalitis in fish,” Curr.Microbiol.New York, vol.28, pp.139-143, March 1 994. [8] E.R.Moxon, J.S.Kroll, “The role of bacterial polysaccharide capsules as virulence factors,” Curr.Top.Microbiol. Immu- nol.Berlin, vol.150, pp.65-85, May 199 0. [9] A.S.Cross,“The biologic significance of bacterial encapsula- tion, ” Curr.Top. Microbiol. Immunol. Berlin , vol.150, pp.87-95, May 1990. [10] D.O.Chaffin, S.B.Beres, H.H.Yim, C.E.Rubens, “The serotype of type Ia and III group B streptococci is determined by the poly- merase gene within the polycistronic capsule operon,” J.Bacteriol.Washington DC, vol.182, pp.4466-4477, May 2000. [11] E.Persson, S.Berg, B.Trollfors, P.Larsson, E.Ek, E.Backhaus, B.E.B.Claesson, L.Jonsson, G.Radberg, T.Ripa, S.Johansson, “Serotypes and clinical manifestations of invasive group B streptococcal infections in western Sweden 1998–2001,” Clin.Microbiol.Infect.Wiley, New York, vol.10, pp.791-796, September 2004. [12] H.C.Slotved, F.Kong, L.Lambertsen, S.Sauer, G.L.Gilbert, “Se- rotype IX, a proposed new Streptococcus agalactiae se r o type ,” J. Clin. Microbiol. Washington DC, vol.45, pp.2929-2936, Sep- tember 2007. [13] M.E.Hickman, M.A.Rench, P.Ferrieri, C.J.Baker, “Changing epid emiology of group B strep tococcal coloni zation,” Pediatrics. American Academy of Pediatrics, New York, vol.104, pp.203-209, August 1999. [14] M.Waqner, T.Murai, B.Waqner, E.Gunther, J.Jelinkova, “JM9 strains, a new type of group B streptococci from Japan,” Zen- tra lbl.Bakteriol. Elsevier, Amsterdam, vol.280, pp.488-496, Marc h 1994.  X. FU ET AL. Copyright © 2012 SciRes. ENG 171 [15] N.Suanyuk, F.Kong, D.Ko, G.L.Gilbert, K.Supamattaya, “Oc- currence of rare genotypes of Streptococcus agalactiae in cul- tu red red ti lapia Oreochromis sp. and Nile tila pia O. niloticus in Thailand—Relationship to human isolates?, ”Aquaculture, El- sevier , Amst erd a m, vol.284, pp.35-40, July 2008. [16] P.Vandamme, L.A.Devriese, B.Pot, K.Kersters, P.Melin, “Streptococcus difficile is a nonhemolytic group B, type Ib Streptococcus,” IJSEM. Society for General Microbiology, London , vol.47, pp.81-85, January 199 7. [17] F.Stingele, J.R.Neeser, B.Mollet, “Identification and characteri- zati on of the eps (Exopolysac charide) gene cluster from St re p to- coccus thermophilus Sfi6,” J.Bacteriol. Washington DC, vol.178, pp.1680-1690, March 1996. [18] S.Yamamoto, K.Miyake, Y.Koike, M.Watanabe, Y.Machida, M.Ohta, S.Iijima, “Molecular characterization of type-specific capsular polysaccharide biosynthesis genes of Streptococcus aga la ctiae type Ia, ”J.Bacteriol. Washington DC, vol.181, pp.5176-5184, S ep tember 1999. [19] A.Guidolin, J.K.Morona, R.Morona, D.Hansman, J.C.Paton, “Nucleotide sequence analysis of genes essential for capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 19F,” Infect. Immun. Washington DC, vol.62, pp.5384-5396, December 1994. [20] M.J.Cieslewicz, D.L.Kasper, Y.Wang, M.R.Wessels, “Function- al analysis in type Ia group B Streptococcus of a clu ster of g enes involved in extracellular polysaccharide production by diverse species of streptococci,” J. Biol. Chem. Washington DC, vol.276, pp.139-146, J anuary 2001 . [21] J.K.Morona, J.C.Paton, D.C.Miller, R.Morona, “Tyrosine phos- phorylation of CpsD negatively regulates capsular polysaccha- ride biosynthesis in Streptococcus pneumoniae,” Mol. Micro- bio l.Wiley, New York, vol.35, pp.1431-1442, March 20 0 0. [22] C.E.Rubens, L.M.Heggen, R.F.Haft, M.R.Wessels, “Identifica- tion of cpsD,a gene essential for type III capsule expression in group B streptococc i,” Mol. Mic robiol. Wiley, New York, vol.8, pp.843-855, May 1993. [23] A. M. Baya, B. Lupiani, F. M. Hetrick, B. S. Roberson, R. Lu- kacovic, E. May, C. Poukish, “Association of Streptococcus sp. with fish mortalities in the Chesapeake Bay and its tributaries,” J.Fish.Dis.Wiley,New York, vol.13, p p.251-253, May 1990. [24] R.Duremdez, A. A.Marzouk, J. A. Qasem, A. A.Harbi, H. Gha- rabally, “ Isolati on of Streptococcus agalactiae from cultured sil- ver pomfret, Pampus argenteus (Euphrasen), in Kuwait,” J.Fish.Dis.Wiley,New York, vol.27, pp.307-310, May 2004. [25] N.Suanyuk, H.Kanghear, R. Khongpradit, K.Supamattaya, “Streptococcus agalactiae infecti on in ti lap ia (Oreochromis nilo- ticus),” J. Sci. Technol. Aquat .Sci. Songkhla, vol.27, pp.307-319, 2005. [26] M. Imperi, M. Pataracchia, G. Alfarone, L. Baldassarri, G. Ore- fici, R. Creti, “A multiplex PCR assay for the direct identifica- tion of th e capsular t ype (Ia to IX) of Streptococcus agalactiae,” J. Microbiol. Meth ods, Els evi er, Ams t erd a m, v ol. 80, pp.212-214, December 2009. [27] D. O. Chaffin, L. M. Mentele, C.E. Rubens, “Sialylation of group B streptococcal capsular polysaccharide is mediated by cp sK and is required for optimal capsule polymerization and ex- pression,” J.Bacteriol. Washington DC, vol.187, pp.4615-4626, July 2005. [28] A. L. Larentis, A. P. C. Argondizzo, G. dos S. Esteves, El. Jes- souron, R. Galler, M. A.Medeiros, “Cloning and optimization of induction conditions for mature PsaA (pneumococcal surface adhesin A) expression in Escherichia coli and recombinant pro- tein stability during long-term storage,” Protein Expr.Purif. El- sevier, Amsterdam, vol .78, pp.38-47, Jul y 2011. |

