 Journal of Cancer Therapy, 2013, 4, 1283-1289 http://dx.doi.org/10.4236/jct.2013.48151 Published Online October 2013 (http://www.scirp.org/journal/jct) 1283 Nuclear Odontogenic Ameloblast-Associated Protein (ODAM) Correlates with Melanoma Sentinel Lymph Node Metastasis Sagar S. Gandhi1,2, Daniel P. Kestler2,3, Charles T. Bruker4, James M. McLaughlin1,2, Robert E. Heidel2, Sabina Siddiqui5, James S. Foster2,3, Keith D. Gray1,2, John Bell1,2, Alan Solomon 2,3, James Lewis1,2 1Department of Surgery, University of Tennessee Medical Center-Knoxville, Knoxville, USA; 2Graduate School of Medicine, Uni- versity of Tennessee Medical Center-Knoxville, Knoxville, USA; 3Department of Medicine, University of Tennessee Medical Cen- ter-Knoxville , Knoxville, USA; 4Department of Pathology, Boca Raton Regional Hospital, Boca Raton, USA; 5Department of Sur- gery, University of Michigan School of Medicine, Ann Arbor, USA. Email: dkestler@utmck.edu Received August 8th, 2013; revised September 2nd, 2013; accepted September 9th, 2013 Copyright © 2013 Sagar S. Gandhi et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT We have examined primary tumor sections from melanoma patients by immunohistochmistry (IHC) for the presence of the odontogenic ameloblast-associated protein (ODAM). Within these patient tissues we have observed a correlation of nuclear ODAM staining in the primary tumors with sentinel lymph node (SLN) metastasis. Surgically, SLN invasion in melanoma is considered an important indicator of more aggressive, invasive melanoma and to date there are limited biomarkers which strongly correlate with metastatic disease. The observation that ODAM staining in melanoma associ- ates with SLN invasion may have important prognostic implications which could assist in the management of mela- noma. Notably, ODAM expression may correlate with pathway-signaling we have previously reported to be affected by ectopic ODAM expression in cultured melanoma and breast cancer cell lines. Keywords: Melanoma; Sentinel Lymph Node Metastases; ODAM; Immunohistochemistry 1. Introduction Melanoma metastasis is predicted by factors that reflect biologic behavior such as primary tumor Breslow thick- ness, mitotic rate, and ulceration [1,2]. Sentinel lymph node (SLN) status in melanoma remains the single most important predictor of overall survival [3-5]. In addition, records from the AJCC Melanoma Staging Database demonstrate that as Breslow thickness increases, a sig- nificant decline in both 5- and 10-year survival rates is observed, and recent data demonstrate a significant cor- relation between survival and the primary tumor mitotic rate. Notably, survival rates of patients with an ulcerated melanoma and similar Breslow thickness are signifi- cantly worse compared to non-ulcerated matched pri- mary tumors [2]. Many potential biomarkers for mela- noma have been reported, but their clinical significance largely remains undetermined [6]. Molecular factors in- fluencing primary melanoma growth and metastasis re- flect dysregulation of normal cellular signaling pathways, and these factors continue to be intensively investigated, both with respect to potential therapeutic advances and for utility as prognostic indicators [7]. ODAM is a protein initially identified as the amyloid- forming component in a rare dental neoplasm, calcifying epithelial odontogenic tumor or Pindborg tumor. The protein has been detected in a broad range of epithelial tissues and in multiple human cancers including those of the breast, esophagus, gastric tissues, and bronchial epi- thelium [8,9]. The potential role of this protein as a marker of disease status and survival in breast cancer has been reported as increased nuclear ODAM staining of primary breast tumors across disease staging [10]. A portion of patients (up to ~13% in late stage) who were ODAM-positive exhibited improved survival compared to stage-matched ODAM-negative breast cancer patients while the remaining bulk (~87%) of late stage ODAM- positive tumor patients did not exhibit a survival benefit, suggesting at least two patients’ outcome groups associ- ated with ODAM-expressing breast tumors [10]. Based on previous observations that lymph node-positive breast Copyright © 2013 SciRes. JCT  Nuclear Odontogenic Ameloblast-Associated Protein (ODAM) Correlates with Melanoma Sentinel Lymph Node Metastasis 1284 cancer patients are often positive for nuclear ODAM staining, together with the propensity of melanomas to metastasize into regional lymph nodes, we examined ODAM expression in primary tumors and lymph node biopsies of patients with SLN-positive (Stage III) and SLN-negative (Stage I-II) melanoma. This allowed us to test whether nuclear ODAM staining in primary mela- noma could predict sentinel node positivity. We report our findings as follows. 2. Materials and Methods 2.1. Melanoma Patient Tumor Tissue Patients diagnosed with melanoma were identified retro- spectively through our institutional tumor registry from years 2000-2006. Only cases with available primary tu- mor and SLN tissue were evaluated. Histological features of the primary tumors were recorded for both the SLN- negative and positive samples, along with overall sur- vival (OS) and recurrence data obtained from our patient database. Archived formalin-fixed paraffin embedded tu- mor tissues were cut and immunostained with murine monoclonal anti-ODAM antibody 8B4, as previously reported [10] and detailed below. 2.2. IHC Analysis Two micrometer thick formalin-fixed paraffin embedded melanoma sections were mounted on charged slides (Fisherbrand Superfrost/Plus, Thermo-Fisher), dried over- night at room temperature, and deparaffinized to water. Sections were immersed in antigen-retrieval solution (Biogenex Citra Plus, BioGenex, San Ramon, CA, USA) and subjected to standard blocking procedures. Anti- ODAM antibody was applied at 1:7500 in diluent (Dako, #S-3022), incubated overnight (5˚C), and visualized us- ing the ImmPRESS polymerized enzyme-linked reporter system followed by the ImmPACT diaminobenzidine de- tection kit (Vector Laboratories, Burlingame, CA, USA). External positive controls were utilized for slide inter- pretation with each batch of patient tumor-slides while, benign structures present in each study section served as internal controls. Tumor tissue obtained from each block was stained by hematoxylin-eosin and reviewed in conjunction with im- munostained slides. The presence or absence of ODAM immunostaining was determined in the neoplastic cells of each case and staining was assessed and reported, for the nucleus only, as negative or positive. Nuclear positivity was defined by the presence of distinct smooth homoge- nous staining of at least 50% of tumor cell nuclei, and negativity was defined as a near complete lack of nuclear immunostaining in essentially all tumor cells of interest (less than 1% of nuclei) as in previous studies [10]. This was based on the observed distribution of nuclear ODAM staining where positive tumors exhibited staining in essentially all (greater than 90%) cell nuclei while staining was present in less than 1% of cell nuclei in tu- mors designated as ODAM-negative. Thus, tumors with a percent reactivity between 1% and 90% were rare, and 50% reactivity was chosen as a cutoff to provide a study design that minimized ambiguity. In practice, no tumors exhibited reactivity near the 50%-positive cutoff. Stain- ing for ODAM was graded in blinded fashion by a single peer reviewed pathologist (CTB). 2.3. Statistics Descriptive statistics were conducted for the Breslow thickness, age, gender, and months to follow-up. An in- dependent samples t-test was used to compare Breslow values on SLN-positive and SLN-negative groups. In the event of a violation of a statistical assumption, a non- parametric Mann-Whitney U test was employed. Unad- justed odds ratios (OR) with 95% confidence intervals (CI) were calculated to compare ODAM staining and SLN status to various discrete variables including recur- rence, cancer status, and cancer death. Logistic regres- sion analysis was employed to yield multivariate adjusted odds ratios when predicting for SLN positivity. Statisti- cal significance was assumed at a p < 0.05 level and all analyses were conducted using SPSS Version 19 soft- ware (SPSS, Chicago, IL). 3. Results 3.1. Patient Sample Populations Our institutional tumor registry contained 270 cases of primary melanoma treated from 2000-2006. Complete data and adequate tissue samples were available for 44 of these patients (21 SLN-positive patients and 23 SLN- negative patients). Inadequate tissue samples, incomplete medical records, non-sentinel lymph node biopsies and patients who underwent lymphadenectomies were ex- cluded. All patients had invasive melanoma. Table 1 depicts patient demographics and primary tumor charac- teristics for each cohort. Demographics were not signifi- cantly different but was close to significance in regards to Breslow thickness (p = 0.09). Ulceration was not sig- nificantly different between cohorts, 13 versus 10 in the SLN-positive and SLN-negative cohorts, respectively (p = 0.61). 3.2. ODAM Immunostaining Immunostaining for ODAM showed that both the nu- cleus and cytoplasm of benign melanocytes stain strongly ositive for ODAM, while in melanoma, staining of the p Copyright © 2013 SciRes. JCT  Nuclear Odontogenic Ameloblast-Associated Protein (ODAM) Correlates with Melanoma Sentinel Lymph Node Metastasis Copyright © 2013 SciRes. JCT 1285 Table 1. Melanoma patient characteristicsa. SLN-NEGATIVE SLN-POSITIVE ODAM-NEGATIVE ODAM-POSITIVE AGE (Median) 68 (44 - 85) 54 (34 - 75) 68 (34 - 85) 56 (39 - 80) GENDER (M:F) 10:13 12:9 8:15 14:7 LOCATION Extremities 14 8 13 9 Trunk 4 9 7 6 Head 5 4 3 6 MEDIAN BRESLOW (Range) 2 mm (0.35 - 7) 3.38 mm (0.32 - 16) 2.48 mm (0.35 - 10) 2.75 mm (0.32 - 16) ULCERATION PRESENT 10 13 13 10 aCohort Demographics are separated by sentinel lymph node-negative (Stage I/II), sentinel lymph node-positive (Stage III), ODAM-negative, and ODAM- positive melanoma. cytoplasm is consistently diminished and nuclear staining is variable. In ODAM-negative melanoma there is no nuclear staining of the primary tumor (Figure 1), while ODAM-positive melanoma exhibits readily demonstrable staining in the cell nucleus (Figure 2). Primary tumors in the SLN-positive cohort were significantly more likely to exhibit nuclear localization of ODAM as recorded in Table 2. Sixteen of 21 specimens (76%) in the SLN- positive patients stained for ODAM compared to 5 of 23 (22%) in the SLN-negative patients (Odds Ratio (OR) = 11.52, 95% CI 2.81, 47.23). SLN staining for ODAM corresponded with the primary tissue staining pattern in all specimens. No SLN stained positive for ODAM unless the primary tumor was ODAM-positive as well. Our study had 13 thin melanomas (≤1 mm), 5 of which (38%) were ultimately Stage III. Notably, of these five, four stained positive for ODAM. Also, 4 of 13 thin melanomas were ulcerated; 1 of these was SLN-negative, 3 were SLN-positive, and ODAM status correlated with SLN status in all samples from ulcerative tumors. 3.3. Recurrence and Survival Analyses Median follow-up for the SLN-positive and negative cohorts was 37 months (range of 7 - 68) and 47 months (range of 4 - 64), respectively. Four patients were lost to follow-up, all of which were in the SLN-negative group (half were ODAM-positive). Logistic regression analysis, given in Table 3, found that when controlled for Breslow thickness and ulceration, participants that stained ODAM- positive were 35 times more likely to be SLN-positive (OR = 35, 95% CI 4.05 - 302.26). Breslow thickness was close to showing a significant association (OR 1.48, 95% CI 0.96 - 2.27, p = 0.077). As shown in Table 4, disease recurrence developed in 10 of 18 (55.5%) node positive patients and 2 of 22 (9.1%) node negative patients (OR = 12.5, 95% CI 2.23, 70.19). Three patients in the SLN-positive group were found to have metastatic disease during staging after ini- tial wide local excision and SLN biopsy and were thus considered never disease free. Subset analysis of ODAM- positive patients irrespective of nodal status demon- strated that 10 of 20 (50%) ODAM-positive patients had recurrence versus 2 of 20 (10%) in the ODAM-negative group (OR = 9, 95% CI 1.64, 49.45). Overall survival (OS) was 10 of 21 (48%) in the SLN-positive group and 18 of 23 (79%) in the SLN-negative group (3.96, 95% CI 1.07, 14.67). OS in the ODAM-positive group was 11 of 21 (52%) versus 17 of 23 (74%) for ODAM-negative primary tumors (OR = 0.39, 95% CI 0.11, 1.38) as dis- played in Table 5. 4. Discussion Our study revealed that SLN-positive primary tumors (Stage III) were significantly more likely to exhibit nu- clear ODAM upon IHC staining, and thus suggests that nuclear ODAM localization is associated with more in- vasive tumors. Furthermore, 4 of 5 SLN-positive thin melanomas (<1 mm) were found to be nuclear ODAM- positive. Since an estimated 5% - 8% of thin melanomas metastasize to lymph nodes, SLN biopsies are not rou- tinely performed in these cases [5]. We propose that preoperative staining of melanoma, and particularly thin melanoma, for ODAM may guide operative management and patient treatment. ODAM is expressed in a broad range of normal epi- thelial tissues and malignancies [8,9]. During dental de- velopment the protein is secreted from ameloblasts and associated with the junctional epithelium at the incisor enamel interface [11,12]. Late in dental development ODAM localizes to the ameloblast nucleus and mediates direct activation of MMP-20/enamelysin gene expression [13]. Thus, while its roles have not been fully delineated,  Nuclear Odontogenic Ameloblast-Associated Protein (ODAM) Correlates with Melanoma Sentinel Lymph Node Metastasis 1286 Figure 1. ODAM-negative primary melanoma (stage I). Left: hematoxylin-eosin stain. Right: anti-ODAM immunostain. Arrows indicate tumor cell nuclei. Note brown staining of phagocytic cells with lack of nuclear staining in tumor cells. Original magnifications 200×. Figure 2. ODAM-positive primary melanoma (stage III). Left: hematoxylin-eosin stain (original magnification 100×). Right: anti-ODAM immunostain (original magnification 200×). The arrows indicate two of numerous ODAM-posi- tive tumor cell nuclei. ODAM exhibits potential cell signaling functions both in the nucleus and through interactions with extracellular matrix components, suggesting classification of ODAM as a matricellular protein [12-14]. Proteins of this subset include the tenascin, osteopontin, thrombospondin, SPARC (osteonectin), SPARCL1 (Hevin), and CCN proteins. This class of proteins has been suggested to contribute to melanoma progression by supporting cellular release from keratinocyte control [15]. The high degree of cellular organization of normal dif- ferentiated tissues is often lost in cancer. The detection of ODAM nuclear localization in melanoma is consistent with the observation of other proteins, such as activating transcription factor 2 (ATF2), showing increased local- ization to the nucleus in metastatic melanoma [16]. Nu- clear re-localization of forkhead box 03 (FOXO3a), β- catenin, and a number of other proteins is proving to be a determining factor in tumor cell growth and invasiveness associated with a broad range of malignancies including melanoma [17-19]. As of yet, no singular molecular biomarkers have pro- ven clinical utility for predicting the progression of mela- noma to metastasis [20-22]. Previous studies have dis- cussed biomarkers such as S100B and lactate dehydro- genase as prognostic indicators for melanomas [6,20-24]. These biomarkers are elevated in advanced disease, and their presence indicates poor prognosis and diminished survival. However, they are not routinely used in clinical practice for early stage disease. Similarly, markers for lymphatic vessel density have been utilized as predictors of SLN metastasis, but this requires analysis of multiple antigens by IHC and a high degree of variability has been reported [25]. Recent reports have also described two potential biomarkers for melanoma identified by mono- clonal antibodies KBAb2 and PNL2 and observed in over 85% of cases but the associated antigens have not, to date, been identified [26]. A previous retrospective study of breast cancer tumor sections at our institution demonstrated a statistically sig- nificant correlation between the presence of nuclear ODAM and tumor stage [10]. Our current study also demon- strates that ODAM expression correlates with melanoma recurrence, survival, and that Breslow thickness approach- ed significance when associated with ODAM staining. These observations underlie recent research in our laboratory which demonstrated profound growth inhibi- tion in vitro and in vivo with human breast cancer cells engineered to express ODAM. This corresponds with suppression of the PI3K/AKT/mTOR signaling pathway [27,28]. Notably, melanoma exhibits frequent dysregula- tion of the PI3K/AKT/mTOR and RAS/RAF/MAPK sig- naling pathways [7,29,30] and we have observed growth suppression in vitro and AKT inhibition upon ectopic ODAM expression in human melanoma cell lines [28]. This suggests potential impacts on tumor cell behavior, disease progression, and therapeutic outcomes when ODAM is expressed in these malignancies. Yet, in the presence of functional, or dysregulated signaling path- way components, these effects may differ. Our studies thus serve to highlight the complex interactions between signaling pathways in melanoma, given that single drug inhibitors can yield contrary effects on tumor behavior dependent upon the cellular context [31-34]. Recognizing the complexity of ODAM function with respect to cellular localization, and the participation of multiple signaling pathways in driving melanoma growth, clarification of the role of ODAM expression in mela- noma biology is necessary both in mechanistic terms, and as support for any possible clinical utility of staining for ODAM. The study described herein was of moderate size and retrospective in nature. Thus, the correlation of ODAM expression with SLN positivity and overall survival will be better delineated by analyses comprised of greater sample size and longer follow-up intervals, which will allow for increased statistical power in the results. Further, a clear as- sessment of the association of ODAM with melanoma spread will also depend on studies which place ODAM ex- pression in the context of evolving subtype classifications for melanoma [35] and the associated molecular hallmarks of these subtypes (e.g.; BRAF V600E, c-KIT, NRAS, cy- clin D). Copyright © 2013 SciRes. JCT 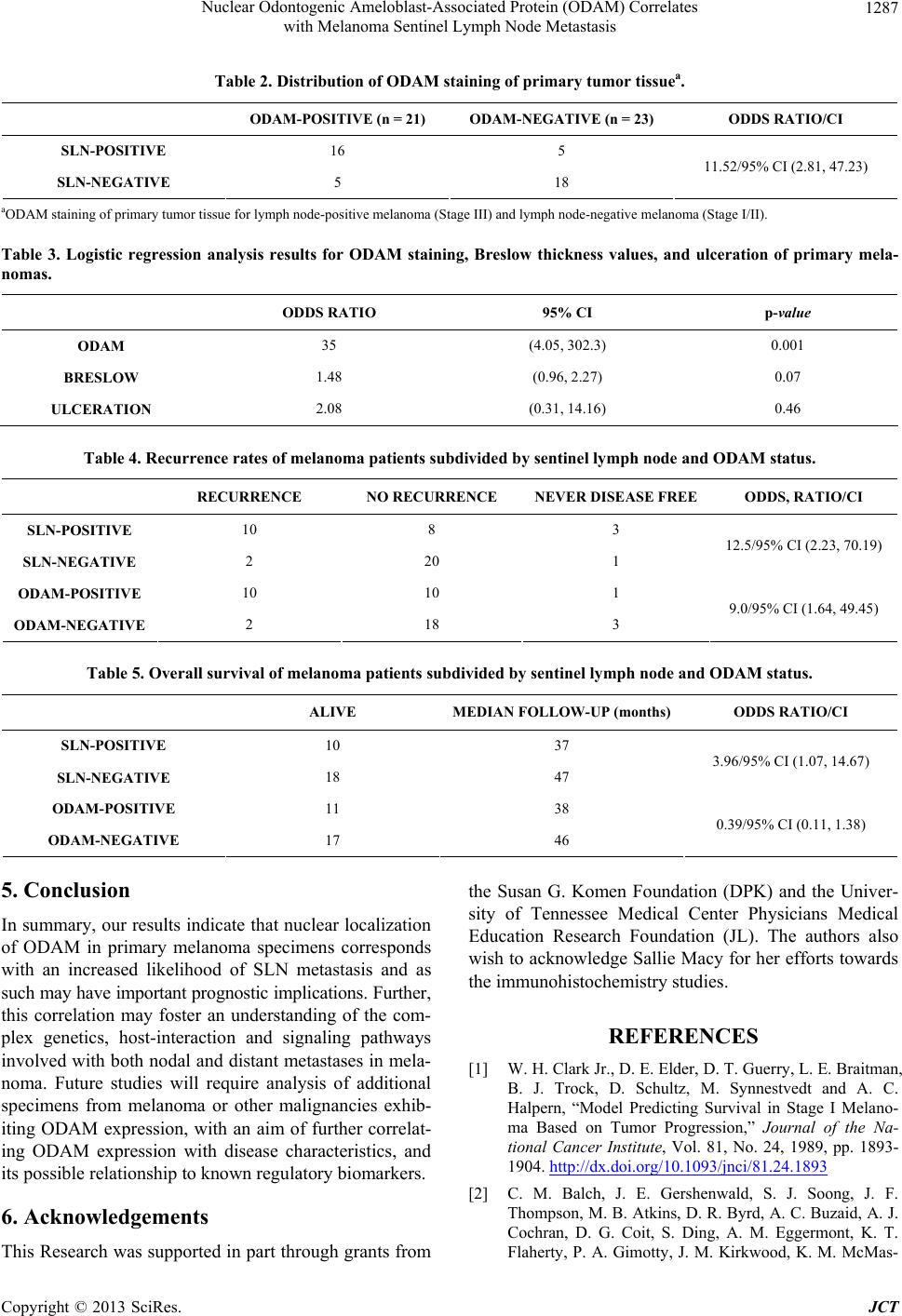 Nuclear Odontogenic Ameloblast-Associated Protein (ODAM) Correlates with Melanoma Sentinel Lymph Node Metastasis Copyright © 2013 SciRes. JCT 1287 Table 2. Distribution of ODAM staining of primary tumor tissuea. ODAM-POSITIVE (n = 21) ODAM-NEGATIVE (n = 23) ODDS RATIO/CI SLN-POSITIVE 16 5 SLN-NEGATIVE 5 18 11.52/95% CI (2.81, 47.23) aODAM staining of primary tumor tissue for lymph node-positive melanoma (Stage III) and lymph node-negative melanoma (Stage I/II). Table 3. Logistic regression analysis results for ODAM staining, Breslow thickness values, and ulceration of primary mela- nomas. ODDS RATIO 95% CI p-value ODAM 35 (4.05, 302.3) 0.001 BRESLOW 1.48 (0.96, 2.27) 0.07 ULCERATION 2.08 (0.31, 14.16) 0.46 Table 4. Recurrence rates of melanoma patients subdivided by sentinel ly mph node and ODAM status. RECURRENCE NO RECURREN CE NEVER DISEASE FREE ODDS, RATIO/CI SLN-POSITIVE 10 8 3 SLN-NEGATIVE 2 20 1 12.5/95% CI (2.23, 70.19) ODAM-POSITIVE 10 10 1 ODAM-NEGATIVE 2 18 3 9.0/95% CI (1.64, 49.45) Table 5. Overall survival of melanoma patients subdivided by sentine l lymph node and ODAM status. ALIVE MEDIAN FOLLOW-UP (months)ODDS RATIO/CI SLN-POSITIVE 10 37 SLN-NEGATIVE 18 47 3.96/95% CI (1.07, 14.67) ODAM-POSITIVE 11 38 ODAM-NEGATIVE 17 46 0.39/95% CI (0.11, 1.38) 5. Conclusion In summary, our results indicate that nuclear localization of ODAM in primary melanoma specimens corresponds with an increased likelihood of SLN metastasis and as such may have important prognostic implications. Further, this correlation may foster an understanding of the com- plex genetics, host-interaction and signaling pathways involved with both nodal and distant metastases in mela- noma. Future studies will require analysis of additional specimens from melanoma or other malignancies exhib- iting ODAM expression, with an aim of further correlat- ing ODAM expression with disease characteristics, and its possible relationship to known regulatory biomarkers. 6. Acknowledgements This Research was supported in part through grants from the Susan G. Komen Foundation (DPK) and the Univer- sity of Tennessee Medical Center Physicians Medical Education Research Foundation (JL). The authors also wish to acknowledge Sallie Macy for her efforts towards the immunohistochemistry studies. REFERENCES [1] W. H. Clark Jr., D. E. Elder, D. T. Guerry, L. E. Braitman, B. J. Trock, D. Schultz, M. Synnestvedt and A. C. Halpern, “Model Predicting Survival in Stage I Melano- ma Based on Tumor Progression,” Journal of the Na- tional Cancer Institute, Vol. 81, No. 24, 1989, pp. 1893- 1904. http://dx.doi.org/10.1093/jnci/81.24.1893 [2] C. M. Balch, J. E. Gershenwald, S. J. Soong, J. F. Thompson, M. B. Atkins, D. R. Byrd, A. C. Buzaid, A. J. Cochran, D. G. Coit, S. Ding, A. M. Eggermont, K. T. Flaherty, P. A. Gimotty, J. M. Kirkwood, K. M. McMas-  Nuclear Odontogenic Ameloblast-Associated Protein (ODAM) Correlates with Melanoma Sentinel Lymph Node Metastasis 1288 ters, M. C. Mihm Jr., D. L. Morton, M. I. Ross, A. J. So- ber and V. K. Sondak, “Final Version of 2009 AJCC Melanoma Staging and Classification,” Journal of Clini- cal Oncology, Vol. 27, No. 36, 2009, pp. 6199-6206. http://dx.doi.org/10.1200/JCO.2009.23.4799 [3] F. Amersi and D. L. Morton, “The Role of Sentinel Lymph Node Biopsy in the Management of Melanoma,” Advances in Surgery, Vol. 41, 2007, pp. 241-256. http://dx.doi.org/10.1016/j.yasu.2007.05.015 [4] D. L. Rousseau, Jr., M. I. Ross, M. M. Johnson, V. G. Prieto, J. E. Lee, P. F. Mansfield and J. E. Gershenwald, “Revised American Joint Committee on Cancer Staging Criteria Accurately Predict Sentinel Lymph Node Positiv- ity in Clinically Node-Negative Melanoma Patients,” An- nals of Surgical Oncology, Vol. 10, No. 5, 2003, pp. 569- 574. http://dx.doi.org/10.1245/ASO.2003.09.016 [5] D. L. Morton, J. F. Thompson, R. Essner, R. Elashoff, S. L. Stern, O. E. Nieweg, D. F. Roses, C. P. Karakousis, N. Mozzillo, D. Reintgen, H. J. Wang, E. C. Glass and A. J. Cochran, “Validation of the Accuracy of Intraoperative Lymphatic Mapping and Sentinel Lymphadenectomy for Early-Stage Melanoma: A Multicenter Trial. Multicenter Selective Lymphadenectomy Trial Group,” Annals of Surgery, Vol. 230, No. 4, 1999, pp. 453-463, Discussion 463-455. [6] B. E. Gould Rothberg, M. B. Bracken and D. L. Rimm, “Tissue Biomarkers for Prognosis in Cutaneous Mela- noma: A Systematic Review and Meta-Analysis,” Journal of the National Cancer Institute, Vol. 101, No. 7, 2009, pp. 452-474. http://dx.doi.org/10.1093/jnci/djp038 [7] I. Yajima, M. Y. Kumasaka, N. D. Thang, Y. Goto, K. Takeda, O. Yamanoshita, M. Iida, N. Ohgami, H. Tamura, Y. Kawamoto and M. Kato, “RAS/RAF/MEK/ERK and PI3K/PTEN/AKT Signaling in Malignant Melanoma Pro- gression and Therapy,” Dermatology Research and Prac- tice, Vol. 2012, 2012, Article ID: 354191. http://dx.doi.org/10.1155/2012/354191 [8] A. Solomon, C. L. Murphy, K. Weaver, D. T. Weiss, R. Hrncic, M. Eulitz, R. L. Donnell, K. Sletten, G. Wester- mark and P. Westermark, “Calcifying Epithelial Odonto- genic (Pindborg) Tumor-Associated Amyloid Consists of a Novel Human Protein,” Journal of Laboratory and Clinical Medicine, Vol. 142, No. 5, 2003, pp. 348-355. http://dx.doi.org/10.1016/S0022-2143(03)00149-5 [9] D. P. Kestler, J. S. Foster, S. D. Macy, C. L. Murphy, D. T. Weiss and A. Solomon, “Expression of Odontogenic Ameloblast-Associated Protein (ODAM) in Dental and Other Epithelial Neoplasms,” Molecular Medicine, Vol. 14, No. 5-6, 2008, pp. 318-326. http://dx.doi.org/10.2119/2008-00010.Kestler [10] S. Siddiqui, C. T. Bruker, D. P. Kestler, J. S. Foster, K. D. Gray, A. Solomon and J. L. Bell, “Odontogenic Amelo- blast Associated Protein as a Novel Biomarker for Human Breast Cancer,” The American Surgeon, Vol. 75, No. 9, 2009, pp. 769-775. [11] J. C. Park, J. T. Park, H. H. Son, H. J. Kim, M. J. Jeong, C. S. Lee, R. Dey and M. I. Cho, “The Amyloid Protein APin is Highly Expressed during Enamel Mineralization and Maturation in Rat Incisors,” European Journal of Oral Sciences, Vol. 115, No. 2, 2007, pp. 153-160. http://dx.doi.org/10.1111/j.1600-0722.2007.00435.x [12] P. Moffatt, C. E. Smith, R. St-Arnaud and A. Nanci, “Characterization of Apin, a Secreted Protein Highly Ex- pressed in Tooth-Associated Epithelia,” Journal of Cellu- lar Biochemistry, Vol. 103, No. 3, 2008, pp. 941-956. http://dx.doi.org/10.1002/jcb.21465 [13] H. K. Lee, D. S. Lee, H. M. Ryoo, J. T. Park, S. J. Park, H. S. Bae, M. I. Cho and J. C. Park, “The Odontogenic Ameloblast-Associated Protein (ODAM) Cooperates with RUNX2 and Modulates Enamel Mineralization via Regu- lation of MMP-20,” Journal of Cellular Biochemistry, Vol. 111, No. 3, 2010, pp. 755-767. http://dx.doi.org/10.1002/jcb.22766 [14] C. Nishio, R. Wazen, S. Kuroda, P. Moffatt and A. Nanci, “Expression Pattern of Odontogenic Ameloblast-Asso- ciated and Amelotin during Formation and Regeneration of the Junctional Epithelium,” European Cells and Mate- rials, Vol. 20, 2010, pp. 393-402. [15] M. Fukunaga-Kalabis, A. Santiago-Walker and M. Herlyn, “Matricellular Proteins Produced by Melanocytes and Melanomas: In Search for Functions,” Cancer Microen- vironment, Vol. 1, No. 1, 2008, pp. 93-102. http://dx.doi.org/10.1007/s12307-008-0009-0 [16] A. J. Berger, H. M. Kluger, N. Li, E. Kielhorn, R. Hala- ban, Z. Ronai and D. L. Rimm, “Subcellular Localization of Activating Transcription Factor 2 in Melanoma Speci- mens Predicts Patient Survival,” Cancer Research, Vol. 63, No. 23, 2003, pp. 8103-8107. [17] R. G. Bedolla, Y. Wang, A. Asuncion, K. Chamie, S. Siddiqui, M. M. Mudryj, T. J. Prihoda, J. Siddiqui, A. M. Chinnaiyan, R. Mehra, R. W. de Vere White and P. M. Ghosh, “Nuclear versus Cytoplasmic Localization of Filamin A in Prostate Cancer: Immunohistochemical Cor- relation with Metastases,” Clinical Cancer Research, Vol. 15, No. 3, 2009, pp. 788-796. http://dx.doi.org/10.1158/1078-0432.CCR-08-1402 [18] J. J. Derry, G. S. Prins, V. Ray and A. L. Tyner, “Altered Localization and Activity of the Intracellular Tyrosine Kinase BRK/Sik in Prostate Tumor Cells,” Oncogene, Vol. 22, No. 27, 2003, pp. 4212-4220. http://dx.doi.org/10.1038/sj.onc.1206465 [19] S. P. Tenbaum, P. Ordonez-Moran, I. Puig, I. Chicote, O. Arques, S. Landolfi, Y. Fernandez, J. R. Herance, J. D. Gispert, L. Mendizabal, S. Aguilar, S. Ramon y Cajal, S. Schwartz, Jr., A. Vivancos, E. Espin, S. Rojas, J. Baselga, J. Tabernero, A. Munoz and H. G. Palmer, “Beta-Catenin Confers Resistance to PI3K and AKT Inhibitors and Sub- verts FOXO3a to Promote Metastasis in Colon Cancer,” Nature Medicine, Vol. 18, No. 6, 2012, pp. 892-901. http://dx.doi.org/10.1038/nm.2772 [20] N. K. Haass and K. S. Smalley, “Melanoma Biomarkers: Current Status and Utility in Diagnosis, Prognosis, and Response to Therapy,” Molecular Diagnosis and Therapy, Vol. 13, No. 5, 2009, pp. 283-296. http://dx.doi.org/10.1007/BF03256334 [21] A. R. Larson, E. Konat and R. M. Alani, “Melanoma Biomarkers: Current Status and Vision for the Future,” Nature Clinical Practice Oncology, Vol. 6, No. 2, 2009, Copyright © 2013 SciRes. JCT  Nuclear Odontogenic Ameloblast-Associated Protein (ODAM) Correlates with Melanoma Sentinel Lymph Node Metastasis Copyright © 2013 SciRes. JCT 1289 pp. 105-117. http://dx.doi.org/10.1038/ncponc1296 [22] C. W. Joyce, I. G. Murphy, M. Rafferty, D. Ryan, E. W. McDermott and W. M. Gallagher, “Tumor Profiling Us- ing Protein Biomarker Panels in Malignant Melanoma: Application of Tissue Microarrays and Beyond,” Expert Review of Proteomics, Vol. 9, No. 4, 2012, pp. 415-423. http://dx.doi.org/10.1586/epr.12.5 [23] T. S. Wang, T. M. Johnson, P. N. Cascade, B. G. Redman, V. K. Sondak and J. L. Schwartz, “Evaluation of Staging Chest Radiographs and Serum Lactate Dehydrogenase for Localized Melanoma,” Journal of the American Academy of Dermatology, Vol. 51, No. 3, 2004, pp. 399-405. http://dx.doi.org/10.1016/j.jaad.2004.02.017 [24] U. Bottoni, P. Izzo, A. Richetta, T. J. Mannooranparampil, V. Devirgiliis, M. Del Giudice, M. G. Reale, L. Frati and S. Calvieri, “S100 Serum Level: A Tumour Marker for Metastatic Melanoma,” Melanoma Research, Vol. 13, No. 4, 2003, pp. 427-429. http://dx.doi.org/10.1097/00008390-200308000-00014 [25] S. Pasquali, A. P. van der Ploeg, S. Mocellin, J. R. Stretch, J. F. Thompson and R. A. Scolyer, “Lymphatic Biomar- kers in Primary Melanomas as Predictors of Regional Lymph Node Metastasis and Patient Outcomes,” Pigment Cell and Melanoma Research, Vol. 26, No. 3, 2013, pp. 326-337. http://dx.doi.org/10.1111/pcmr.12064 [26] P. P. Aung, M. Sarlomo-Rikala, J. Lasota, J. P. Lai, Z. F. Wang and M. Miettinen, “KBA62 and PNL2: 2 New Melanoma Markers-Immunohistochemical Analysis of 1563 Tumors Including Metastatic, Desmoplastic, and Mucosal Melanomas and Their Mimics,” American Jour- nal of Surgical Pathology, Vol. 36, No. 2, 2012, pp. 265- 272. http://dx.doi.org/10.1097/PAS.0b013e31823651cb [27] D. P. Kestler, J. S. Foster, C. T. Bruker, J. W. Prenshaw, S. J. Kennel, J. S. Wall, D. T. Weiss and A. Solomon, “ODAM Expression Inhibits Human Breast Cancer Tu- morigenesis,” Breast Cancer: Basic and Clinical Re- search (Auckl), Vol. 5, 2011, pp. 73-85. [28] J. S. Foster, L. M. Fish, J. E. Phipps, C. T. Bruker, J. M. Lewis, J. L. Bell, A. Solomon and D. P. Kestler, “Odon- togenic Ameloblast-Associated Protein (ODAM) Inhibits Growth and Migration of Human Melanoma Cells and Elicits PTEN Elevation and INACTIVATION of PI3K/ AKT Signaling,” BMC Cancer, Vol. 13, 2013, p. 227. http://dx.doi.org/10.1186/1471-2407-13-227 [29] J. A. Curtin, J. Fridlyand, T. Kageshita, H. N. Patel, K. J. Busam, H. Kutzner, K. H. Cho, S. Aiba, E. B. Brocker, P. E. LeBoit, D. Pinkel and B. C. Bastian, “Distinct Sets of Genetic Alterations in Melanoma,” New England Journal of Medicine, Vol. 353, No. 20, 2005, pp. 2135-2147. http://dx.doi.org/10.1056/NEJMoa050092 [30] H. Tsao, V. Goel, H. Wu, G. Yang and F. G. Haluska, “Genetic Interaction between NRAS and BRAF Muta- tions and PTEN/MMAC1 Inactivation in Melanoma,” Journal of Investigative Dermatology, Vol. 122, No. 2, 2004, pp. 337-341. http://dx.doi.org/10.1046/j.0022-202X.2004.22243.x [31] L. P. Cantini, F. Meier, V. K. Sondak and K. S. Smalley, “The Future of Targeted Therapy Approaches in Mela- noma,” Expert Opinion on Drug Discovery, Vol. 4, No. 4, 2009, pp. 445-456. http://dx.doi.org/10.1517/17460440902828298 [32] W. H. Chappell, L. S. Steelman, J. M. Long, R. C. Kempf, S. L. Abrams, R. A. Franklin, J. Basecke, F. Stivala, M. Donia, P. Fagone, G. Malaponte, M. C. Mazzarino, F. Nicoletti, M. Libra, D. Maksimovic-Ivanic, S. Mijatovic, G. Montalto, M. Cervello, P. Laidler, M. Milella, A. Ta- furi, A. Bonati, C. Evangelisti, L. Cocco, A. M. Martelli and J. A. McCubrey, “Ras/Raf/MEK/ERK and PI3K/ PTEN/Akt/mTOR Inhibitors: Rationale and Importance to Inhibiting These Pathways in Human Health,” Onco- target, Vol. 2, No. 3, 2011, pp. 135-164. [33] J. B. Korman and D. E. Fisher, “Developing Melanoma Therapeutics: Overview and Update,” Wiley Interdisci- plinary Reviews: Systems Biology and Medicine, Vol. 5, No. 3, 2013, pp. 257-271. http://dx.doi.org/10.1002/wsbm.1210 [34] T. Shimizu, A. W. Tolcher, K. P. Papadopoulos, M. Bee- ram, D. W. Rasco, L. S. Smith, S. Gunn, L. Smetzer, T. A. Mays, B. Kaiser, M. J. Wick, C. Alvarez, A. Cavazos, G. L. Mangold and A. Patnaik, “The Clinical Effect of the Dual-Targeting Strategy Involving PI3K/AKT/ mTOR and RAS/MEK/ERK Pathways in Patients with Advanced Cancer,” Clinical Cancer Research, Vol. 18, No. 8, 2012, pp. 2316-2325. http://dx.doi.org/10.1158/1078-0432.CCR-11-2381 [35] S. J. Vidwans, K. T. Flaherty, D. E. Fisher, J. M. Tenen- baum, M. D. Travers and J. Shrager, “A Melanoma Mo- lecular Disease Model,” PLoS One, Vol. 6, No. 3, 2011, Article ID: e18257. http://dx.doi.org/10.1371/journal.pone.0018257
|