 Vol.4, No.9, 483-490 (2013) Agricultural Sciences http://dx.doi.org/10.4236/as.2013.49065 RNAi of MiASB caused high mortality of Meloidogyne incognita juveniles and inhibited the nematode disease Yonghong Huang1,2*, Mei Mei1,3*, Baoming Shen1,3, Zhenchuan Mao1, Bingyan Xie1# 1Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China; #Corresponding Author: xiebingyan@caas.cn, albertluke@126.com 2Key Laboratory of Biology and Genetic Resource Utilization of Fruit Tree in South Subtropics, Ministry of Agriculture/Fruit Research Institute, Guangdong Academy of Agricultural Science, Guangzhou, China 3College of Horticulture and Landscape, Hunan Agriculture University, Changsha, China Received 27 April 2013; revised 28 May 2013; accepted 10 June 2013 Copyright © 2013 Yonghong Huang et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT The southern root-knot nematode, Meloidogyne incognita, is one of the most prevalent and damaging plant-parasitic nematodes in the world and causes serious damages to agricul- tural production. W e cloned a mitoc hondrial ATP synthase b subunit gene fragment of M. incog- nita (MiASB) based on the nematode genomics prediction. By soaking in the MiASB dsRNA so- lution, the hatching of RNAi treated eggs was reduced by 60% compared to negative control and by 64% compared to untreated control. Mortality of RNAi treated second stage juvenile (J2) was 8.6 times higher than that of negative control and 26 times higher than the untreated control. Inoculating the RNAi treated egg mass- es and J2 to tomato seedlings showed the pa- thogencity was significantly reduced. For the RNAi treated egg masses, the amount of root galls on silence treated seedlings was reduced by 92% compared to that on the negative control seedlings, and reduced by 93% compared to that on untreated control seedlings. For the treated J2, the amount of root galls on silence treated seedlings was reduced by 83% and 86% com- pared to negative and untreated control seed- lings, respectively. The study revealed the Mi- ASB silence had a positive effect on prevention and control of root-knot nematode disease, and also showed that the MiASB may be involved in the pathogenesis of nematode, which provided new ideas and ways to the researc h of nematode pathology and nematode disease control. Keywords: Root-knot Nematode; Meloidogyne Spp.; Mitochondrial ATP Synthase; RNA Interference; dsRNA 1. INTRODUCTION Root-knot nematodes (RKN), genus Meloidogyne, are root endoparasites that can develop on a wide range of plant species. The infective second stage juvenile (J2) penetrates the root tip and migrates intercellularly until it reaches the differentiating vascular cylinder [1]. It in- duces root-knot, stunts nutrient deficiency and disrupts the physiology of the host plants through their reproduc- tion and feeding within plant roots, leading to a signify- cant reduction in crop yield and deterioration of product quality [2,3]. Crop infestation by RKN causes 70 billion U.S. dollars of crop losses annually in fruit and vegetable production [4]. The RKN is being controlled in the fields mainly based on cultural practices, host-plant resistance and the application of synthetic pesticides. Nematode control has become difficult in recent years owning to the withdrawal or restricted use of effective synthetic pesticides for environmental problems and human and animal health concerns [3,5-7], which urges the need for alternative control methods. Strategies based on the spe- cific blocking of parasitism gene products involving in the success of infection would offer attractive alterna- tives to reduce nematode populations in the field [8]. RNA interference (RNAi) is a mechanism for post- transcriptional gene silencing. This technique uses the fact that exposure of an organism to double-stranded RNA (dsRNA) from a gene of interest causes posttrans- criptional silencing of the endogenous gene and allows a null phenotype to be mimicked [9]. Since it was firstly *These authors contribute equally to the article. Copyright © 2013 SciRes. OPEN ACCES S  Y. H. Huang et al. / Agricultural Sciences 4 (2013) 4 83-490 484 discovered over two decade ago in Caenorhabditis ele- gans [10], RNAi have been proved to be a widespread phenomenon shared by many phyla [11]. In C. elegans, targeting the dsRNA into the nematode intestine by microinjection or ingestion allows the silencing of genes expressed in distal tissues. In Nippostrongylus brasilien- sis, dsRNA uptake into the nematode body was induced by the cationic lipid polymer lipofectin. In Brugia malayi, a low-volume culture system was developed to expose females to dsRNA [1]. Nowadays, RNAi has been devel- oped as a powerful tool for studying gene function in a variety of organisms [12]. Mitochondria and chloroplasts serve as power stations of living cells. By generating the biological energy cur- rency ATP, ATP synthases play a decisive role in this process [13]. Mitochondrial ATP synthase (also named F1F0-ATP synthase or complex V) is located in the inner mitochondrial membrane together with respiratory chain complexes I-IV [14] and is a rotating nanomotor in pro- karyotic and eukaryotic cells that uses a transmembrane electrochemical ion gradient as an energy source to con- vert ADP and Pi to ATP [15]. F1F0-ATP synthase con- sists of up to 18 different subunits [16], each of which is the essential components for ATP synthase to generate ATP. In preliminary studies, based on the genomic data of C. elegans, M. hapla, M. incognita and RNAi data in Wormbase, we predicted some functional genes in Meloi- dogyne nematode, one of which was ASB encoding the b subunit of the F0 proton channel portion of F1F0-ATP synthase. In this study, we cloned a 650 bp ASB fragment from M. incognita (MiASB). Using RNAi by soaking it in the dsRNA solution of MiASB, we investigated the ef- fects of MiASB on the hatching of the egg masses and the mortality of Meloidogyne nematode juveniles, and also studied its inhibitory effect on nematode disease caused by M. incognita. 2. MATERIALS AND METHODS 2.1. Nematode Collection Nematode M. incognita was extracted from the pure culture that was previously initiated by egg masses and propagated on tomato (Solanum lycopersicum) in the glasshouse. Egg masses were handpicked using sterilized forceps from heavily infected roots 45 days after incuba- tion. The egg masses were surface-sterilized (1% NaOCl) for 1 min followed by washing with sterile distilled water. Some of the egg masses was placed on 15 mesh sieves (8 cm in diameter) containing crossed layers of tissue paper in Petri-dishes with sterile distilled water just deep enough to contact the egg masses and incubated 25˚C to 26˚C to obtain freshly hatched J2. Emerged J2 were col- lected daily and stored at 4˚C. J2 up to 4 days old were used in experiments [17]. 2.2. Plant Material Tomato (Solanum lycopersicum cv. Lichun) was used in the experiment. Seeds of tomato were soaked in warm water for 6 h, then were placed on the sterile filter paper in a Petri dish for germinating at 25˚C in dark. The ger- minated seeds were sown in pots containing culture sub- strate composed of peat and vermiculite at 2 to 1 ratio in greenhouse. Seedlings with 3 - 4 true leaves were used for further experiments. 2.3. Molecular Cloning of MiASB Fragment Total RNA was isolated from M. incognita J2 using Trizol (Invitrogen), following the manufacturer’s proto- col. The first strand was synthesized using SuperScript TM III First-Strand Synthesis System for RT-PCR (In- vitrogen) according to manufacturer’s instructions. A 650 bp fragment of MiASB was amplified by PCR using Mi- ASB-specific primers including MiASB-F (5’-TGT TGG ACA AAC TTT TGC TT-3’) and MiASB-R (5’-AAA TTT TCT TGG GTT GCT TTG-3’) designed according to the MiASB (GeneBank Accession: JQ247982), a gene we cloned in the preliminary studies based on the pre- dicted MiASB according to the nematode genomics using bioinformatics methods. Polymerase chain reaction (PCR) was performed using 2 μl of the first-strand reaction as a template. Amplifica- tions were performed with Taq DNA polymerase (2.5 U, Promega) using 1 mM of each of the primers and 200 mM of each of the deoxy nucleotide triphosphates (dNTP). The conditions were as follows: 94˚C, 3 min; followed by 35 cycles of 94˚C, 30 s; 53˚C, 30 s; 72˚C, 60 sand a final cycle of 72˚C, 10 min. The resulting PCR products were analyzed by agarose gel electrophoresis. Bands with the correct size were excised from the gel, purified using TaKaRa MiniBEST Agarose Gel DNA Extraction Kit Ver.3.0 and cloned into pGM-T Vector (TA Cloning Kit, Tiangen) according to the manufac- turer’s instructions, forming the vector pGM-MiASB. The pGM-MiASB was transformed into the competent cells of E. coli DH5α and incubated at 37˚C overnight. The white clones grown on the LB medium containing Ampicil- lin/X-gal/IPTG were picked and the plasmids were ex- tracted and sent to Invitrogen (Beijing) Co., Ltd., Beijing, China for sequencing. 2.4. Plasmid Recombinant and in Vitro Transcription to Produce dsRNA The pGM-MiASB was digested with restriction en- donuclease Eco RI (TaKaRa), the resulting products of which were analyzed by agarose gel electrophoresis, bands with the small size were excised from the gel, pu- Copyright © 2013 SciRes. OPEN ACCES S  Y. H. Huang et al. / Agricultural Sciences 4 (2013) 4 83-490 485 rified using TaKaRa MiniBEST Agarose Gel DNA Ex- traction Kit Ver.3.0, ligated to the LITMUS 28 i plasmid (NEB, USA)digested with the exactly same restriction endonuclease using T4 DNA ligase (Invitrogen), result- ing in the plasmid LITMUS-MiASB. and then trans- formed into competent cells of E. Coli DH5α and incu- bated at 37˚C overnight. LITMUS-MiASB plasmid from a single positive colony was confirmed by sequencing. To insert T7 RNA polymerase site into the two ends of the MiASB, 1 μl of the identified LITMUS-MiASB plas- mid were used as a template to perform another PCR. Amplifications were performed with Taq DNA poly- merase (2.5 U, Promega) using 2 mM of T7 primer (5’-TAA TAC GAC TCA CTA TAG G-3’) and 10 mM of each of the deoxy nucleotide triphosphates (dNTP), with an initial denaturation step of 94˚C for 5 min, followed by 35 cycles of 94˚C for 30 s, 55˚C for 30 s, 72˚C for 50 s, and a final elongation step of 72˚C for 10 min. After amplification, the PCR products (T7-MiASB-T7) were separated by electrophoresis on agarose gel and purified using TaKaRa MiniBEST Agarose Gel DNA Extraction Kit Ver.3.0. Then the purified T7-MiASB-T7 fragment was used to generatedds RNAs using in vitro Transcrip- tion T7 Kit (TaKaRa) following the manufacturer’s in- structions. The synthesized dsRNAs was stored at −70˚C for further experiments. 2.5. RNAi of MiASB by Soaking RNAi experiment was performed in a 24-microwell plate according to the previous studies [1,9,18] with slight modification. Uniform single egg mass or ap- proximately 2000 freshly J2 were deposited in each well and socked in M9 buffer solution (43.6 mM Na2HPO4, 22 mM KH2PO4, 18.7 mM NH4Cl, and 8.6 mM NaCl) containing 1% resorcinol, 50 mM octopamine, 3 mM spermidine, 0.05% gelatin, and dsRNA at 2 mg/ml in the dark at room temperature on a rotator (RNAi treatment). One control samples were incubated in the same solution but without dsRNA (Negative control). And the other control samples were incubated in the sterile water and also no dsRNA was added (Untreated control). For the egg RNAi treatment, half of the egg masses was treated just for 1 day and the total cumulative number of juve- nile hatched from the egg masses were counted one days later (1-day-treatment), and the other half of the egg masses were treated for 3 days and total cumulative number of juvenile from these egg masses were counted 3 days later (3-day-treatment) to estimate the RNAi ef- fect on the hatching of the egg masses. The cumulative numbers of dead juvenile in 1-day- and 3-day-treatments were also recorded at the corresponding time point, re- spectively, to essay the RNAi effect on the mortality of juveniles. For the J2 RNAi treatment, Total J2 and the dead J2 was counted 6 h after the treatmentto determine RNAi effect on the mortality of J2. The juvenile that was straight and did not move even after mechanical prod- ding were defined as dead. 2.6. Infection of Plants The RNAi treated egg masses (1-day-treatment and 3-day-treatment) and RNAi treated J2 were applied close to the roots of seedlings using a sterilized micropiette. 3 RNAi treated egg masses or approximately 6000 RNAi treated J2 were inoculated to each seedling. The two corresponding controls (negative control and untreated control) were also inoculated according to the RNAi treatment. These treated seedlings were cultured in the greenhouse. The seedlings were uprooted 45 days later, and root galls were recorded to evaluate the effects of the MiASB RNAi on the disease caused by M. incognita. 3. RESULTS 3.1. Generation of Mi A SB dsRNA A 650 bp fragment of MiASB was obtained by PCR using total RNA of M. incognita J2 as template. The identity of the cloned MiASB fragment with the corre- sponding parts of the MiASB (GeneBank) accession: JQ247982) was as high as 100%.To generate the dsRNA of MiASB (Figure 1), the recombinant pLITMUS-Mi- ASB was firstly constructed by linearizing pGM-MiASB and pLITMUS-28i with Eco RI, respectively, and ligated with T4 DNA ligase. Then pLITMUS-MiASB were used to performed PCR as a template with T7 Primer and the T7 polymerase site was inserted into two ends of the MiASB and formed the fragment T7-MiASB-T7. After that, in vitro transcription was catalyzed by T7 poly- merase with fragment T7-MiASB-T7 as template andds RNA of MiASB was finally generated. 3.2. MiASB RNAi Inhibited Hatching of M. incognita Egg Masses On the first day, juveniles were hatched from the egg masses, the amounts of which were slightly but not sig- nificantly different (F = 0.98, P = 0.3975). On the third- days, the cumulative juvenilea mounts in the RNAi treatment was significantly lower than those in negative and untreated controls (F = 14.9, P = 0.003). The average amountin MiASB treatment was 63, which was reduced by 60% and 64% compared to negative and untreated controls, respectively (Figure 2). The juvenile amount innegative control was slightly, but not significantly, higher than untreated control treatment, which showed the reagents supplemented in the RNAi solution hardly influenced the hatching of the egg masses, but RNAi of MiASB significantly inhibited the hatching of egg masses Copyright © 2013 SciRes. OPEN ACCES S 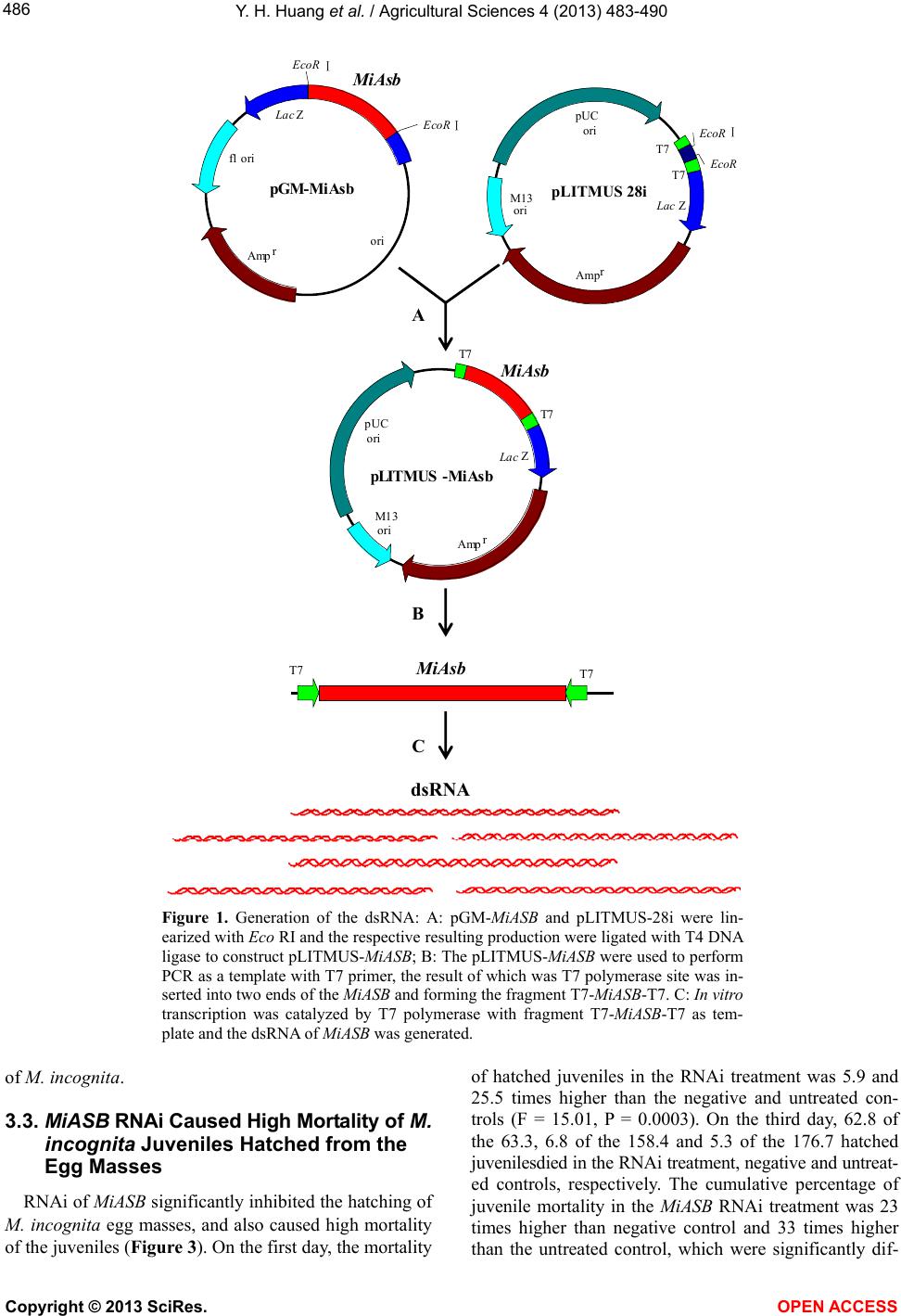 Y. H. Huang et al. / Agricultural Sciences 4 (2013) 4 83-490 Copyright © 2013 SciRes. 486 pLITMUS -MiA sb 5000bp La c Amp M13 orir T7 T7 Z ori pUC Mi Asb pGM-MiAsb 4237bp La c MiAsb Amp ori f1 ori r Ⅰ Ⅰ ZEcoR EcoR MiAsb T7 T7 dsRNA pLITMUS 28i 4237bp Lac Amp M13 ori r Ⅰ Z ori pUC T7 T7 EcoR EcoR A B C Figure 1. Generation of the dsRNA: A: pGM-MiASB and pLITMUS-28i were lin- earized with Eco RI and the respective resulting production were ligated with T4 DNA ligase to construct pLITMUS-MiASB; B: The pLITMUS-MiASB were used to perform PCR as a template with T7 primer, the result of which was T7 polymerase site was in- serted into two ends of the MiASB and forming the fragment T7-MiASB-T7. C: In vitro transcription was catalyzed by T7 polymerase with fragment T7-MiASB-T7 as tem- plate and the dsRNA of MiASB was generated. of hatched juveniles in the RNAi treatment was 5.9 and 25.5 times higher than the negative and untreated con- trols (F = 15.01, P = 0.0003). On the third day, 62.8 of the 63.3, 6.8 of the 158.4 and 5.3 of the 176.7 hatched juvenilesdied in the RNAi treatment, negative and untreat- ed controls, respectively. The cumulative percentage of juvenile mortality in the MiASB RNAi treatment was 23 times higher than negative control and 33 times higher than the untreated control, which were significantly dif- of M. incognita. 3.3. MiASB RNAi Caused High Mortality of M. incognita Juveniles Hatched from the Egg Masses RNAi of MiASB significantly inhibited the hatching of M. incognita egg masses, and also caused high mortality of the juveniles (Figure 3). On the first day, the mortality OPEN A CCESS  Y. H. Huang et al. / Agricultural Sciences 4 (2013) 4 83-490 487 ferent (F = 6417.04, P < 0.0001). 3.4. MiASB RNAi Caused High Mortality of M. incognita J2 M. incognita J2 was soaked in the dsRNA solution di- rectly and were died. 6 h after treatment, about 434 of the 2000 J2 was found dead in the RNAi treatment, but 50 of the 2000 J2 in the negative control and 17 of 2000 J2 in untreated control were dead, respectively. The J2 mortal- ity of RNAi treatment was 8.6 and 26 times higher than the negative and untreated controls, respectively (Figure 4), and were significantly different (F = 157.34, P < 0.0001). 3.5. MiASB RNAi Treated Egg Masses Reduced Root Galls on Tomato Seedlings RNAi treated egg masses were inoculated to the to- Figure 2. The number of juveniles hatched from MiASB RNAi treated egg masses of M. incognita was significantly lower than those hatched from the negative and untreated controls 3 days later. Figure 3. The mortality of juveniles hatched from MiASB RNAi treated egg masses was significantly higher than those hatched from the negative and untreated controls. mato seedlings. 45 days later, the root galls were induced on all the tomato seedlings. For the 1-day-treatment, the number of root galls were between 18 and 30 in the RNAi treated seedlings, and those ranged from 48 to 112 for the two controls. The average number of root galls on RNAi treated seedling sreduced by 70.6% and 74.7% compared to the negative and untreated control seedlings, respectively (Figure 5) and were significantly different (F = 37.1, P < 0.0001). For the 3-day-treatment, the root gall amounts on the RNAi treated seedling were between 0 and 10, but those on the two controls ranged from 30 to 79. The average number of root galls on negative control and untreated control seedlings were 51.7 and 59, re- spectively, and that on the silenced seedlings was only 3.8 (Figure 5). Statistical analysis showed a significant difference existed among the number of root galls on the three differently treated seedlings (F = 38.1, P < 0.0001). The amount of root galls on the RNAi treated seedlings reduced by 92% and 93% compared to the negative and untreated control seedlings, respectively. Figure 4. The mortality of J2 in the MiASB RNAi treatment was significantly higher than those of the negative and un- treated controls 6 h later. Figure 5. The root galls induced on the tomato seedlings in- oculated with MiASB RNAi treated egg masses were signifi- cantly reduced compared to those inoculated with the negative and untreated control egg masses 45 days later. Copyright © 2013 SciRes. OPEN ACCES S 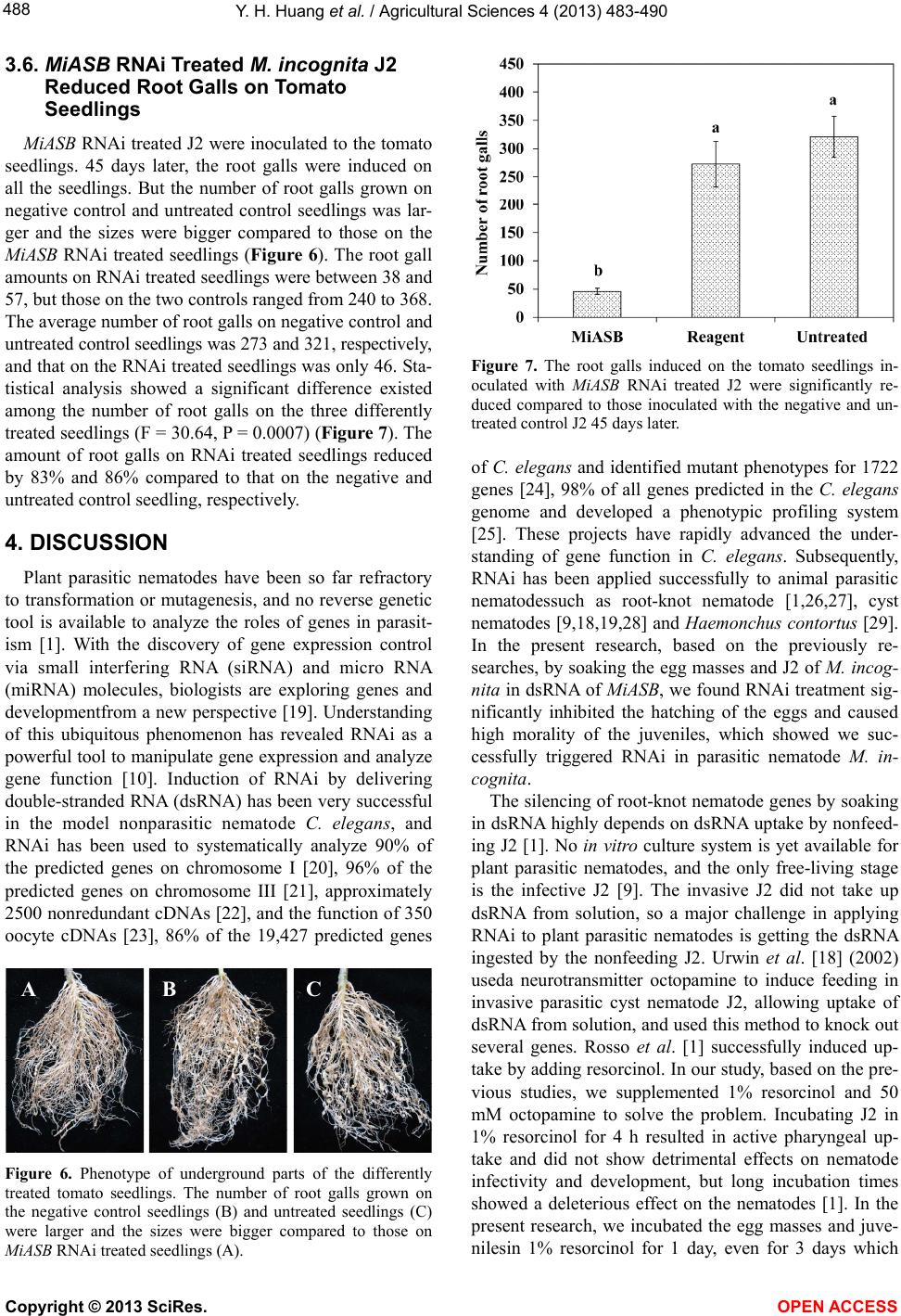 Y. H. Huang et al. / Agricultural Sciences 4 (2013) 4 83-490 488 3.6. MiASB RNAi Treated M. incognita J2 Reduced Root Galls on Tomato Seedlings MiASB RNAi treated J2 were inoculated to the tomato seedlings. 45 days later, the root galls were induced on all the seedlings. But the number of root galls grown on negative control and untreated control seedlings was lar- ger and the sizes were bigger compared to those on the MiASB RNAi treated seedlings (Figure 6). The root gall amounts on RNAi treated seedlings were between 38 and 57, but those on the two controls ranged from 240 to 368. The average number of root galls on negative control and untreated control seedlings was 273 and 321, respectively, and that on the RNAi treated seedlings was only 46. Sta- tistical analysis showed a significant difference existed among the number of root galls on the three differently treated seedlings (F = 30.64, P = 0.0007) (Figure 7). The amount of root galls on RNAi treated seedlings reduced by 83% and 86% compared to that on the negative and untreated control seedling, respectively. 4. DISCUSSION Plant parasitic nematodes have been so far refractory to transformation or mutagenesis, and no reverse genetic tool is available to analyze the roles of genes in parasit- ism [1]. With the discovery of gene expression control via small interfering RNA (siRNA) and micro RNA (miRNA) molecules, biologists are exploring genes and developmentfrom a new perspective [19]. Understanding of this ubiquitous phenomenon has revealed RNAi as a powerful tool to manipulate gene expression and analyze gene function [10]. Induction of RNAi by delivering double-stranded RNA (dsRNA) has been very successful in the model nonparasitic nematode C. elegans, and RNAi has been used to systematically analyze 90% of the predicted genes on chromosome I [20], 96% of the predicted genes on chromosome III [21], approximately 2500 nonredundant cDNAs [22], and the function of 350 oocyte cDNAs [23], 86% of the 19,427 predicted genes A B C Figure 6. Phenotype of underground parts of the differently treated tomato seedlings. The number of root galls grown on the negative control seedlings (B) and untreated seedlings (C) were larger and the sizes were bigger compared to those on MiASB RNAi treated seedlings (A). Figure 7. The root galls induced on the tomato seedlings in- oculated with MiASB RNAi treated J2 were significantly re- duced compared to those inoculated with the negative and un- treated control J2 45 days later. of C. elegans and identified mutant phenotypes for 1722 genes [24], 98% of all genes predicted in the C. elegans genome and developed a phenotypic profiling system [25]. These projects have rapidly advanced the under- standing of gene function in C. elegans. Subsequently, RNAi has been applied successfully to animal parasitic nematodessuch as root-knot nematode [1,26,27], cyst nematodes [9,18,19,28] and Haemonchus contortus [29]. In the present research, based on the previously re- searches, by soaking the egg masses and J2 of M. incog- nita in dsRNA of MiASB, we found RNAi treatment sig- nificantly inhibited the hatching of the eggs and caused high morality of the juveniles, which showed we suc- cessfully triggered RNAi in parasitic nematode M. in- cognita. The silencing of root-knot nematode genes by soaking in dsRNA highly depends on dsRNA uptake by nonfeed- ing J2 [1]. No in vitro culture system is yet available for plant parasitic nematodes, and the only free-living stage is the infective J2 [9]. The invasive J2 did not take up dsRNA from solution, so a major challenge in applying RNAi to plant parasitic nematodes is getting the dsRNA ingested by the nonfeeding J2. Urwin et al. [18] (2002) useda neurotransmitter octopamine to induce feeding in invasive parasitic cyst nematode J2, allowing uptake of dsRNA from solution, and used this method to knock out several genes. Rosso et al. [1] successfully induced up- take by adding resorcinol. In our study, based on the pre- vious studies, we supplemented 1% resorcinol and 50 mM octopamine to solve the problem. Incubating J2 in 1% resorcinol for 4 h resulted in active pharyngeal up- take and did not show detrimental effects on nematode infectivity and development, but long incubation times showed a deleterious effect on the nematodes [1]. In the present research, we incubated the egg masses and juve- nilesin 1% resorcinol for 1 day, even for 3 days which Copyright © 2013 SciRes. OPEN ACCES S  Y. H. Huang et al. / Agricultural Sciences 4 (2013) 4 83-490 489 were far longer than 4 h. To avoid the experiment devia- tion and improve experiment accuracy, we designed two controls, one was negative control to counteract the ef- fect of the reagent including resorcinol and other ele- ments supplemented in the solution. The other was sterile water to replace M9 buffer solution and the supplements. But the results showed no significant difference in the hatching inhibition and juveniles mortality between the two controls. In F1F0-ATP synthase, the static peripheral stalk (δb2) do not appear to be a part of the main stalk, but it has been suggested that they form a second connection, a stator that fixes the catalytic α3β3 subcomplex to the subunit to allow rotation of central stalk and c-ring (cn) [30]. And synthesis of ATP depends upon rotation of the central stalk (γε) and c-ring (cn) [15]. In the present re- search, the nematode egg masses and juveniles were soaked in the dsRNA solution of MiASB, which triggered RNAi in the eggs and the juvenile bodies, resulting in the inhibition of egg hatching and high morality of juveniles, and further reduced the pathogencity. In the process of growth and development, nematodes need ATP as energy to perform various activities including infection. Once the RNAi of MiASB was triggered in the nematode, it caused the reduction of b subunit in ATP synthases, fur- ther impaired the formation of proton channel portion of ATP synthase, finally impacted the production of ATP. In return, the reduced ATP production slowed down the metabolism of the nematode, leading to the low hatching ability, high mortality and reduced infection, which fi- nally resulted in the low nematode disease. 5. CONCLUSION In conclusion, we successfully triggered RNAi in the M. incognita, and RNAi of MiASB significantly reduced the nematode disease. The finding of this research sug- gested MiASB may be associated with the pathogencity of the nematode, and it provided new ideas and ways to further study on nematodes pathogenic mechanisms and prevention and control of nematodes. 6. ACKNOWLEDGEMENTS The research was supported by the Major State Basic Research De- velopment Program of China (Grant No.2009CB119000), Special Fund for Agro-scientific Research in the Public Interest (Grant No. 201103018) and National Natural Science Foundation of China (Grant No. 31030057). We sincerely thank anonymous peer reviewers for the coming suggestions. REFERENCES [1] Rosso, M.N., Dubrana, M.P., Cimbolini, N., Jaubert, S. and Abad, P. (2005) Application of RNA interference to root-knot nematode genes encoding esophageal gland proteins. Molecular Plant-Microbe Interactions, 18, 615- 620. doi:10.1094/MPMI-18-0615 [2] Dang, Q.L., Kim, W.K., Nguyen, C.M., Choi, Y.H., Choi, G.J., Jang, K.S., Park, M.S, Lim, C.H., Lu, N.H. and Kim, J. (2011) Nematicidal and antifungal activities of An- nonaceous acetogeninsfrom Annona squamosa against various plant pathogens. Journal of Agricultural and Food Chemistry, 59, 11160-11167. doi:10.1021/jf203017f [3] Naz, I., Palomares-Rius, J.E., Saifullah, B.V., Khan, M.R., Ali, S. and Ali, S. (2012) In vitro and in plantanematicidal activity of Fumaria parviflora (Fumariaceae) against the southern root-knot nematode Meloidogyne incognita. Plant Pathology, 62, 943-952. doi:10.1111/j.1365-3059.2012.02682.x [4] Caboni, P., Ntalli, N.G., Aissani, N., Cavoski, I. and An- gioni, A. (2012) Nematicidal activity of (E,E)-2,4-Deca- dienal and (E)-2-Decenal from Ailanthus altissima against Meloidogyne javanica. Journal of Agricultural and Food Chemistry, 60, 1146-1151. doi:10.1021/jf2044586 [5] Oka, Y., Shuker, S. and Tkachi, N. (2009) Nematicidal efficacy of MCW-2, a new nematicide of the fluoroal- kenyl group, against the root-knot nematode Meloidogyne javanica. Pest Management Science, 65, 1082-1089. doi:10.1002/ps.1796 [6] Oka, Y., Nacar, S., Putievsky, E., Ravid, U., Yaniv, Z. and Spiegel, Y. (2000) Nematicidal activity of essential oils and their components against the root-knot nematode. Phytopathology, 90, 710-715. doi:10.1094/PHYTO.2000.90.7.710 [7] Ntalli, N.G., Manconi, F., Leonti, M., Maxia, A. and Caboni, P. (2011) Aliphatic ketones from ruta chalepensis (rutaceae) induce paralysis on root knot nematodes. Jour- nal of Agricultural and Food Chemistry, 59, 7098-7103. doi:10.1021/jf2013474 [8] Arguel, M., Jaouannet, M., Magliano, M., Abad, P. and Rosso, M. (2012) siRNAstrigger efficient silencing of a parasitism gene in plant parasitic root-knot nematodes. Genes, 3, 391-408. doi:10.3390/genes3030391 [9] Chen, Q., Rehman, S., Smant, G. and Jones, J.T. (2005) Functional analysis of pathogencity proteins of the potato cyst nematode Globodera rostochiensis using RNAi. Molecular Plant-Microbe Interactions, 18, 621-625. doi:10.1094/MPMI-18-0621 [10] Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E. and Mello, C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806-811. doi:10.1038/35888 [11] Adamo, A., Woglar, A., Silva, N., Penkner, A., Jantsch, V. and La Volpe, A. (2012) Transgene-mediated cosuppres- sion and RNA interference enhance germ-line apoptosis in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of The United States of America, 109, 3440-3445. doi:10.1073/pnas.1107390109 [12] Chen, J., Zhang, D., Yao, Q., Zhang, J., Dong, X., Tian, H., Chen, J. and Zhang, W. (2010) Feeding-based RNA interference of a trehalose phosphate synthase gene in the brown planthopper, Nilaparvata lugens. Insect Molecular Copyright © 2013 SciRes. OPEN ACCES S  Y. H. Huang et al. / Agricultural Sciences 4 (2013) 4 83-490 Copyright © 2013 SciRes. OPEN ACCES S 490 Biology, 19, 777-786. doi:10.1111/j.1365-2583.2010.01038.x [13] Seelert, H. and Dencher, N.A. (2011) ATP synthase super- assemblies in animals and plants: Two or more are better. Biochimica et Biophysica Acta, 1807, 1185-1197. [14] Wittig, I. and Schägger, H. (2009) Supramolecular orga- nization of ATP synthase and respiratory chain in mito- chondrial membranes. Biochimica et Biophysica Acta, 1787, 672-680. doi:10.1016/j.bbabio.2008.12.016 [15] Bisetto, E., Picotti, P., Giorgio, V., Alverdi, V., Mavelli, I. and Lippe, G. (2008) Functional and stoichiometric analysis of subunit e in bovine heart mitochondrial F0F1 ATP synthase. Journal of Bioenergetics and Biomem- branes, 40, 257-267. doi:10.1007/s10863-008-9183-5 [16] Meyer, B., Wittig, I., Trifilieff, E., Karas, M. and Schäg- ger, H. (2007) Identification of two proteins associated with mammalian ATP synthase. Molecular & Cellular Proteomics, 6, 1690-1699. doi:10.1074/mcp.M700097-MCP200 [17] Li, H.Q., Bai, C.Q., Chu, S.S., Zhou, L., Du, S.S., Li, Z.L. and Liu, Q.Z. (2011) Chemical composition and toxicities of the essential oil derived from Kadsura heteroclita stems against Sitophilus zeamais and Meloidogyne incognita. Journal of Medicinal Plants Research, 5, 4943-4948. [18] Urwin, P.E., Lilley, C.J. and Atkinson, H.J. (2002) Inges- tion of doublestranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Molecular Plant- Microbe Interactions, 15, 747-752. doi:10.1094/MPMI.2002.15.8.747 [19] Sindhu, A.S., Maier, T.R., Mitchum, M.G., Hussey, R.S., Davis, E.L. and Baum, T.J. (2009) Effective and specific in planta RNAi in cyst nematodes: expression interfer- ence of four parasitism genes reduces parasitic success. Journal of Experimental Botany, 60, 315-324. doi:10.1093/jxb/ern289 [20] Fraser, A.G., Kamath, R.S., Zipperlen, P., Martinez- Campos, M., Sohrmann, M. and Ahringer, J. (2000) Func- tional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature , 408, 325-330. doi:10.1038/35042517 [21] Gonczy, P., Echeverri, C., Oegema, K., Coulson, A., Jones, S.J.M., Copley, R.R., Duperon, J., Oegema, J., Brehm, M., Cassin, E., Hannak, E., Kirkham, M., Pichler, S., Flohrs, K., Goessen, A., Leidel, S., Alleaume, A.M., Martin, C., Ozlu, N., Bork, P. and Hyman, A.A. (2000) Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature, 408, 331-336. doi:10.1038/35042526 [22] Maeda, I., Kohara, Y., Yamamoto, M. and Sugimoto, A. (2001) Large-scale analysis of gene function in Caenor- habditis elegans by high-throughput RNAi. Current Bi- ology, 11, 171-176. doi:10.1016/S0960-9822(01)00052-5 [23] Piano, F., Schetter, A.J., Mangone, M., Stein, L. and Kemphues, K.J. (2000) RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Current Biology, 10, 1619-1622. doi:10.1016/S0960-9822(00)00869-1 [24] Kamath, R.S., Fraser, A.G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohr- mann, M., Welchman, D.P., Zipperlen, P. and Ahringer, J. (2003) Systematic functional analysis of the Caenorhab- ditis elegans genome using RNAi. Nature, 421, 231-237. doi:10.1038/nature01278 [25] Sönnichsen, B., Koski, L.B., Walsh, A., Marschall, P., Neumann, B., Brehm, M., Alleaume, A.M., Artelt, J., Bettencourt, P., Cassin, E., Hewitson, M., Holz, C., Khan, M., Lazik, S., Martin, C., Nitzsche, B., Ruer, M., Stam- ford, J., Winzi, M., Heinkel, R., Röder, M., Finell, J., Häntsch, H., Jones, S.J., Jones, M., Piano, F., Gunsalus, K.C., Oegema, K., Gönczy, P., Coulson, A., Hyman, A.A. and Echeverri, C.J. (2005) Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Na- ture, 434, 462-469. doi:10.1038/nature03353 [26] Fairbairn, D.J., Cavallaro, A.S., Bernard, M., Mahalinga- Iyer, J., Graham, M.W. and Botella, J.R. (2007) Host-de- livered RNAi: An effective strategy to silence genes in plant parasitic nematodes. Planta, 226, 1525-1533. doi:10.1007/s00425-007-0588-x [27] Charlton, W.L., Harel, H.Y., Bakhetia, M., Hibbard, J.K., Atkinson, H.J. and Mcpherson, M.J. (2010) Additive ef- fects of plant expressed double-stranded RNAs on root- knot nematode development. International Journal for Parasitology, 40, 855-864. doi:10.1016/j.ijpara.2010.01.003 [28] Sukno, S.A., McCuiston, J., Wong, M.Y., Wang, X., Thon, M.R., Hussey, R., Baum, T. and Davis, E. (2007) Quanti- tative detection of double-stranded RNA-mediated gene silencing of parasitism genes in Heterodera glycines. Journal of Nematology, 39, 145-152. [29] Samarasinghe, B., Knox, D.P. and Britton, C. (2011) Factors affecting susceptibility to RNA interference in Haemonchus contortus and in vivo silencing of an H11 aminopeptidase gene. International Journal for Parasi- tology, 41, 51-59. doi:10.1016/j.ijpara.2010.07.005 [30] Walker, J.E. and Dickson, V.K. (2006) The peripheral stalk of the mitochondrial ATP synthase. Biochimica et Biophysica Acta, 1757, 286-296. doi:10.1016/j.bbabio.2006.01.001
|