Journal of Biophysical Chemistry
Vol.2 No.4(2011), Article ID:8428,9 pages DOI:10.4236/jbpc.2011.24044
1,4-DHP-lipid parameters and rod like micellae
1Latvian Institute of Organic Synthesis, Riga, Latvia;
2Faculty of Chemistry, University of Gdansk, Gdansk, Poland; *Corresponding Author: inta@osi.lv
Received 6 July 2011; revised 14 August 2011; accepted 26 August 2011.
Keywords: Lipid; Gene Transfection; Rod-Like Structure; Worm-Like Micellae; Tubular Micellae; Molecular Dynamics
ABSTRACT
The cationic amphiphilic lipid type compound 1,1’-{[3,5-bis(dodecyloxycarbonyl)-4-phenyl-1,4- dihydropyridin-2,6-diyl]dimethylene} bispyridinium dibromide (1,4-DHP lipid) (charge +2), is a gene transfection agent. The electronic structure of 1,4-DHP lipid molecule was investigated by ab initio quantum mechanics, the charges were derived, and the supramolecular structure formed by 1,4-DHP lipid molecules in water was investigated by means of molecular dynamics simulation, f99 force field, version AMBER 8.0. During the molecular dynamics simulations of 10 ns of MD 1,4-DHP lipid formed a worm-like micellae, which was preserved during the time course of 300 ns of MD simulations. Result was confirmed with the electron microscopy showing extended, rod-like structures as one of the 1,4-DHP lipid supramolecular structures, which are expected to be formed at high 1,4-DHP lipid concentrations.
1. INTRODUCTION
New biotechnological drugs, such as DNA, RNA and proteins, require the nanostructure delivery technologies which would pack a DNA or a protein, move through the membrane and release macromolecules inside the cell [1, 2]. Delivered in such a way, a drug targeted at tumors, acts directly in the proper place, and the toxic effect to the whole organism is substantially reduced. For such nanomedicine approach new, different drug delivery agents are required.
DNA delivery into the cell nucleus can be performed by viral carriers, where the guest-DNA is inserted into a virus, or by cationic polymers, cationic peptide or cationic lipid carriers that cap the DNA with the help of electrostatic interactions. Synthetic cationic lipids [3] become increasingly popular as gene transfection agents due to their minimal toxicity, absence of immunological problems, possible large scale production. A large number of lipids with variations in the hydrophilic and hydrophobic regions were generated. Unfortunately, they still have less efficiency when compared with viral vectors. Recently, we synthesized a novel group of gene transfection agents comprising partially hydrogenated cyclic heterocyclic 1,4-dihydropyridine moiety as active linker. 1,4-DHP derivatives are well-known antihypertensive agents, cardiovascular drugs [4], they have peptidomimetic properties, and their structure has been investigated in our earlier studies [5]. Appropriately decorated with substituents, they may possess bioprotective properties [6]. The cationic amphiphilic lipid type compound 1,1’-{[3,5-bis(dodecyloxycarbonyl)-4-phenyl-1,4-dihydropyridin-2,6-diyl]dimethylene} bispyridinium dibromide (1,4-DHP lipid, charge q = +2) (Figure 1) has gene transfection activity [7,8]. This compound is more active than DOTAP and PEI 25 (liposomal and polymeric gene transfection agents). The sharp increase of the 1,4-DHP cycle N-H acidity (up to pKa ≈ 7 - 8) is due to 2- and 6-pyridiniomethyl groups and conjugated alkoxycarbonyl substituents in positions 3 and 5 of the 1,4-DHP cycle. N-H acidity is the basis for its buffering activity in this novel type of gene transfection agents.
The electronic structure of 1,4-DHP lipid molecule was investigated by ab initio quantum mechanics, and the supramolecular structure formed by 1,4-DHP lipid molecules was investigated by means of molecular dynamics simulation.
2. METHODS
1,4-DHP lipid molecule was minimized by Sybyl, with the Tripos force field in vacuo [9] (Figure 1(a)). As it was expected that the fatty acid tails must be not in the fully extended state (Figure 1(a)) in water or lipid bilayer, but close to each other (Figures 1(b)-(d)), the structure with bent fatty acid tails was chosen as the starting structure for lipid supramolecular structure calculations
 (a) (b) (c) (d)
(a) (b) (c) (d)
Figure 1. 1,4-DHP lipid molecule in a space fill and in the stick models: (a) the low energy conformation in vacuo; (b) starting structure for lipid bilayer, front view; (c) starting structure for lipid bilayer, side view; (d) starting structure for lipid bilayer, top view. Minimised by Sybyl, Tripos force field [9].
and also as the starting structure for ab initio quantum mechanics calculations.
The 1,4-DHP lipid structure was calculated by Restricted Hartree-Fock (RHF) ab initio quantum mechanics with the 6-31G* basis set, by using the program GAMESS [10-13], to obtain the charges for molecular dynamics using the RESP algorithm (molecular electrostatic potential based method using charge restrains for determining atom-centered charges).
72 molecules of 1,4-DHP-lipid initially were put in a periodic lipid bilayer-water box, with 10 Å water over the solute and with a small amount of excessive water on the lipid edges to ensure the mobility of lipid molecules (Figures 2(a)-(b)). There were 72 DHP-lipid molecules, 144 counterions of chlorine ions and 4401 water molecules in the system DHP-lipid-water box. The total number of atoms in the system is 22491.
The 1,4-DHP-lipid-water box were subjected to molecular dynamics, (AMBER 8.0, f99 force field, version 8.0 [14-16], NTP protocol (constant number of particles, constant temperature, constant pressure, the derived parameters for 1,4-DHP-lipid are presented in Supplements Table S2).
The temperature was increased gradually from T = 10 K by step of 10 degrees till 300 K. Calculations were performed for 326212 ps.
Results were proceeded with VMD [17], visualization were made with RASMOL [18].
3. RESULTS AND DISCUSSION
1,4-DHP lipid were calculated by Restricted Hartree-
 (a) (b)
(a) (b)
Figure 2. 1,4-DHP lipid initial state of molecular dynamics (a) front vie; (b) side view.
Fock (RHF) ab initio quantum mechanics, 6-31G* bases set, and the quantum minimized structure are depicted in Figure 3. The quantum mechanics calculations show a shallow boat conformation for 1,4-DHP lipid dihydropyridine moiety (Figure 3(b)). The distance between pyridinium nitrogens is 5.9 Å, and the distance between pyridinium carbon atoms in the fourth position is 8 Å, thus the pyridinium rings are upon narrow angle towards each other (Figure 3(a)). The distances between oxygen of the carbonyl group and pyridinium hydrogen are of 2.17 Å - 2.18 Å, and the distances between oxygen of the carbonyl group and the hydrogen atoms of the methylene is of 2.16 and 2.18 Å denoting that the oxygen of the carbonyl group simultaneously interacts with the both hydrogens (Figure 10(a)). The geometric parameters: bond
 (a) (b)
(a) (b)
Figure 3. Ab intito minimized 1,4-DHP-lipid structure— (a) front view; (b) side view. The dihydropyridine ring is in a boat conformation.
length and valence angles are shown in Figure S1.
The lipid head has a +2 charge. The molecular electrostatic potential surfaces were calculated and used to derive the atom point charges for molecular dynamics with RESP algorithm. The representative charges together with derived Amber 8.0 atom types are shown in Figure 4. The full set of charges and atom types for molecular dynamics are presented in Table S1, the corresponding atom numbering is presented in Figure S2.
Molecular dynamics simulations were started from the temperature T = 10 K and risen gradually till 300 K. The temperature changes are represented of Figure 5(a), showing the stepwise rising of the temperature from T = 10 K till T = 300 K, by step of 10 degrees. The total energy with its components of kinetic and potential energy is represented of Figure 5(b), showing that system become stable at 49 ns.
Also the density becomes stable during 49 ns, and further is oscillating around 1.01 g/cm3 (Figure 5(c)).
On Figures 6(a)-(b), representing the MD snapshot at 14725 ps, T = 60 K, we observe that the system still keeps the initial bilayer organization, (6(a) atom type coloring to represent the positions of the charged head groups), but the fatty acid tails of one bilayer squeeze into another bilayer (6(b), molecules are represented in different colors to show the fatty acid tail movement).
On Figures 6(c)-(d) is represented the MD snapshot at the temperature 220 K, 35696 ps of MD run, Figure 6(c) atom type coloring, shows that system still keeps the initial bilayer organization, on Figure 6(d) the different molecule coloring shows that the fatty acid tails of one bilayer squeeze into another bilayer.
After 35 ns of the MD simulation few lipid molecules turned with their charged heads to the side of the lipid bilayer. It is represented on Figures 6(e)-(f), showing 1,4- DHP-lipid at the temperature 300 K 48849 ps of MD run. At this stage of MD run the lipid heads have turned to the sides of the periodic box (Figure 6(e)) and the lipid molecules continue to squeeze from one lipid layer into the other one (Figure 6(f)). After 100 ns of MD simulations a profound worm-like micelle structure began to form. Figures 6(g)-(h) represents a snapshot at 110673 ps of MD run, T = 300 K, and on Figure 6(g) the system, colored by atom type, shows that the 1,4-DHP-lipid charged heads have turned to the sides of the periodic box, and. at this stage of MD run the worm-like micellae has formed. Figure 6(h), where the separate lipid molecules are colored differently shows that molecules from one lipid layer squeeze into the other one; the lipid layers are interwinded.
On Figure 7 is shown the snapshot of 1,4-DHP-lipidwater box at the t = 159582 ps, T = 300 K. The wormlike micelle structure is stable and becomes more perfect

Figure 4. Charge distribution calculated for DHP-lipid molecule and the derived AMBER atom types CA, CM, CT, NA, O, OS, HA, HC, H1, H.
during the course of simulation of 300 ns (Figure 8).
The graphics on Figure 5(d) show that in the supramolecular organization the DHP lipid molecules are in relaxed state, as the bond energy, angle energy and torsion energy increments to the potential energy are small, close to zero, in comparison with electrostatic energy, which is clearly dominating. During the first 50 ns of MD simulations only E-vdw is reduced because of better packing of molecules, while all other components of E_ pot are growing because the system is heated.
The size of the periodic box has changed from the dimensions of x = 70 Å, y = 55 Å, z = 103 Å at the start of molecular dynamics, to x = 102 Å, y = 46 Å, z = 48 Å at 318039 ps (Figure 9). The worm-like micellae axes—the longer dimension, is in the x-axes direction, but y-axes and z-axes dimensions are similar, which is explainable with the top view of the worm-like micellae, having approximately the same diameter in all directions of the yz-plane (Figure 8(b)).
The results of MD simulation were confirmed by electron microscopy, showing the interwinding worm-like structure [19]. The diameter of thinner worm-like structures is around 4 - 5 nm, which corresponds to approximate estimation of the calculated worm-like structure being of 3.8 - 4.5 nm. It could be expected that such worm-

Figure 5. (a) 1,4-DHP lipid molecular dynamics temperature protocol; (b) 1,4-DHP lipid molecular dynamics energy protocol: total energy (red), kinetic energy (blue) and potential energy (green); (c) 1,4-DHP lipid density over the course of molecular dynamics; (d) 1,4-DHP lipid potential energy (red), Van der Waalse energy (green), electrostatic energy (blue), bond energy (violet), angle energy (cyan), torsion energy (yellow) during the course of molecular dynamics.
like structures form on the first stage of lipid molecules association and/or when the number of lipid molecules is relatively high towards the number of water molecules.
It is not surprising that the 1,4-DHP lipid forms structures distinct from a lipid bilayer, as the Restricted Hartree-Fock (RHF) ab initio quantum mechanics calculations show that in the minimized 1,4-DHP lipid structure the lipid head dimension is about 10 Å as the major ellipse axes, and approximately about 8 Å as the minor ellipse axes, if we approximate the lipid head with an ellipse; and the lipid tail width is about 7 Å, so it is so called cone-like lipid prone to form hexagonal assembly [20], tubular structures, which could form a lipoplex [21, 22] and polyplex [23] together with the DNA. The head projection is is πab = 20 Å2, where a and b are one-half of the ellipse’s major and minor axes respectively.
The head-to-tail size ratio of the 1,4-DHP lipid is 10:7 = ~1.43, and this induced certain degree of curvature to the MD starting lamellar the 1,4-DHP lipid system so that they transformed to tubular micellae which could be one of the supramolecular structures in 1,4-DHP lipid self-aggregation.
During the molecular dynamics the both the torsion angles of pyridinium groups and the ester groups of 1,4- DHP-lipid experienced high mobility. On the stage when the tubular micellae is formed, most of the 1,4-DHP lipid molecules tend to turn the pyridinium groups parallely to the micellae’s surface, thus enlarging head to tail size ratio to 15:7 = 2.14. In such a 1,4-DHP lipid molecule conformation the lenght of the molecule is about 21 Å - 22 Å. When the 1,4-DHP lipid molecule’s pyridinium groups are turned parallely to the micellae’s surface, the interactions between the carbonyl oxygen and the hydrogen of the pyridinium group are weakened or lost, when the distances between them enlarged over 3.2 Å, but the interactions between the carbonyl oxygen and the hydrogen of CH2 group still could exist as the distances carbonyl oxygen-CH2 hydrogen are 2.5 Å - 3.07 Å (Figures 10(b) -(d)). The same oxygen-two hydrogens interaction could be realized also by the ester oxygen, pyridinium hydrogen

Figure 6. (a) Snapshot of the 1,4-DHP-lipid-water box at the temperature 60 K, 14725 ps of MD run, space fill representation, colored by atom type; (b) Snapshot of the 1,4-DHP-lipid – water box at the temperature 60 K, 14725 ps of MD run, space fill representation, lipid molecules are colored differently to show that molecules from one lipid layer squeeze into the other one; (c)-(d) snapshot at the temperature 220 K, 35696 ps of MD run; (e)-(f) snapshot at the temperature 300 K 48849 ps of MD run. At this stage of MD run the lipid heads have turned to the sides of the periodic box; (g)-(h) snapshot at the temperature 300 K, 110673 ps of MD run. At this stage of MD run the worm-like micellae has formed.

Figure 7. The snapshot of 1,4-DHP-lipidwater box at the t = 159582 ps of MD run.
 (a) (b)
(a) (b)
Figure 8. 1,4-DHP lipid worm-like micellae is stable during the course of MD simulations. Snapshot at the 300 K, t = 301 ns. (a) Side view; (b) top view.
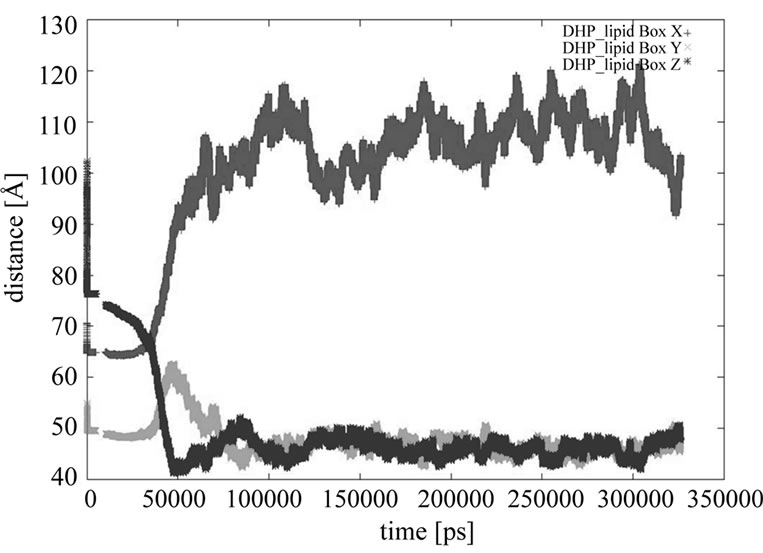
Figure 9. The course of 1,4-DHP lipid periodic box dimensions during the molecular dynamics: x—red, y—green, z—blue. The worm-like micellae axes corresponding to the longer dimension (102 Å), is in the x-axes direction, but the y-axes and z-axes dimensions are similar (46 Å - 48 Å) corresponding to the same diameter of worm-like micellae in the both directions.
 (a)
(a) (b)
(b)
Figure 10. 1,4-DHP lipid molecule ab intio calculations minimization (a) and the snapshots of three 1,4-DHP lipid molecules during at 300 ns of MD (b)-(d).
and CH2 group hydrogen (Figures 10(b)-(d)).
3. CONCLUSIONS
We have performed the Restricted Hartree-Fock (RHF) ab initio quantum mechanics calculations on a 1,4-DHP lipid molecule, as well as 300 ns MD of 72 1,4-DHP lipid molecules starting from a lipid bilayer structure. Results show that the gene transfection agent 1,4-DHP lipid in natural conditions does not form a lipid bilayer, but one of its structures is a worm-like, rod-like micellae, which are expected to be formed at high 1,4-DHP lipid concentrations. We could expect that such the micellaes are capable to form a lipoplex for the DNA transfection. During the molecular dynamics 1,4-DHP lipid molecules tend to turn the piridinium groups parallely to the micellae’s surface.
4. ACKNOWLEDGMENTS
This work was supported by ESF project 2009/0197/1DP/1.1.1.2.0 /09/APIA/VIAA/014, Calculations were performed on computers of the Gdansk Academic Computer Center TASK.
REFERENCES
- Hughes, G.A. (2005) Nanostructure-mediated drug delivery. Nanomedicine: Nanotechnology, Biology, and Medicine, 1, 22-30. doi:10.1016/j.nano.2004.11.009
- Basarkar, A. and Singh, J. (2007) Nanoparticulate systems for polynucleotide delivery. International Journal of Nanomedicine, 2, 353-360.
- Ma, B.-C., Zhang, S.-B., Jiang, H.-M., Zhao, B.-D. and Lv, H.-T. (2007) Lipoplex morphologies and their influences on transfection efficiency in gene delivery. Journal of Controlled Release, 123, 184-194. doi:10.1016/j.jconrel.2007.08.022
- Triggle, D.J. (2006) L-type calcium channels. In: Rampe, D. and Zheng, W., Eds., Voltagegated Ion Channels as Drug Targets, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, 100-122.
- Liepina, I., Blanco, M., Duburs, G. and Liwo, A. (1997) Spatial structure of dihydropyridines and similarity of dihydropyridines with some amino acids. Molecular Engeneering, 7, 401-427. doi:10.1023/A:1008272311650
- Duburs, G., Vigante, B., Plotniece, A., Krauze, A., Sobolevs, A., Briede, J., Klusa, V. and Velena, A. (2008) Dihydropyridine derivatives as bioprotectors. Chemica OGGI/ Chemistry Today, 26, 68-70.
- Hyvonen, Z., Plotniece, A., Reine, I., Chekavichus, B., Duburs, G. and Urtti, A. (2000) Novel cationic amphiphilic 1,4-dihydropyridine derivatives for DNA delivery. Biochimica et Biophysica Acta, 1509, 451-466. doi:10.1016/S0005-2736(00)00327-8
- Hyvonen, Z., Ronkko, S., Toppinen, M.-R., Jaaskelainen, I., Plotniece, A. and Urtti, A. (2004) Dioleoylphosphati Dylethanolamini and PEG-lipid conjugates modify DNA delivery mediated by 1,4-dihydropyridine amphiphiles. Journal of Controlled Release, 99, 177-190. doi:10.1016/j.jconrel.2004.06.019
- SYBYL 8.0, Tripos International, 1699 South Hanley Rd., St. Louis, Missouri, 63144, USA.
- Guest, M.F., Bush, I.J., van Dam, H.J.J., Sherwood, P., Thomas, J.M.H., van Lenthe, J.H., Havenith, R.W.A. and Kendrick, J. (2005) The GAMESS-UK electronic structure package: Algorithms, developments and applications. Molecular Physics, 103, 719-747. doi:10.1080/00268970512331340592
- Zarrabian, S. and Harrison, R.J. (1989) Chemical Physics Letters, 81.
- Saunders, V.R. and van Lenthe, J.H. (1983) Molecular Physics, 48, 923. doi:10.1080/00268978300100661
- Buenker R.J. (1982) Studies in Physical and Theoretical Chemistry, 21, 17.
- Pearlman, D.A., Case, D.A., Caldwell, J.W., Ross, W.S., Cheatham, T.E. III, DeBolt, S., Ferguson, D., Seibel, G. and Kollman, P. (1995) AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Computer Physics Communications, 91, 1-41. doi:10.1016/0010-4655(95)00041-D
- Case, D.A., Cheatham, T., Darden, T., Gohlke, H., Luo, R., Merz, K.M. Jr., Onufriev, A., Simmerling, C., Wang, B. and Woods, R. (2005) The Amber biomolecular simulation programs. Journal of Computational Chemistry, 26, 1668-1688. doi:10.1002/jcc.20290
- Ponder, J.W. and Case, D.A. (2003) Force fields for protein simulations. Advances in Protein Chemistry, 66, 27- 85. doi:10.1016/S0065-3233(03)66002-X
- Humphrey, W., Dalke, A. and Schulten, K. (1996) VMDVisual Molecular Dynamics. Journal of Molecular Graphics, 14, 33-38. doi:10.1016/0263-7855(96)00018-5
- Sayle, R. and Milner-White, E.J. (1995) RasMol: Biomolecular graphics for all. Trends in Biochemical Sciences (TIBS), 20, 374. doi:10.1016/S0968-0004(00)89080-5
- Liepina, I., Czaplewski, C., Ose, V., Danne, R. and Duburs, G. (2008) 1,4-DHP-lipid forms a tubular micellae. In: Hansman. H. E., Meinke, Jan H., Mohanty, S., Nadler W. and Zimmermann, O., Eds., Proceedings of NIC Workshop from Computational Biophysics to System Biology, Julich, 19-21 May 2008, 305-307.
- Gajria, S., Neumann, T. and Tirrell, M. (2011) Self-assembly and applications of nucleic acid solid-state films. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology. Early View, Article First Published Online: 2 June 2011. doi:10.1002/WNAN.148
- Ma, B., Zhang, S., Jiang, H., Zhao, B. and Lv, H. (2007) Lipoplex morphologies and their influences on transfection efficiency in gene delivery. Journal of Controlled Release, 123, 184-194. doi:10.1016/j.jconrel.2007.08.022
- Ulrich, A.S. (2002) Biophysical aspects of using liposomes as delivery vehicles. Bioscience Reports, 22, 129- 150. doi:10.1023/A:1020178304031
- Elouahabi, A. and Ruysschaert, J.-M. (2005) Formation and intracellular trafficking of lipoplexes and polyplexes. Molecular Therapy, 11, 336-347. doi:10.1016/j.ymthe.2004.12.006
Supplements

Figure S1. Geometric parameters: valence bond length (black), valence angles (red) of 1,4-DHP lipid molecule ab initio calculations.

Figure S2. Atom numeration of 1,4-DHP lipid molecule.
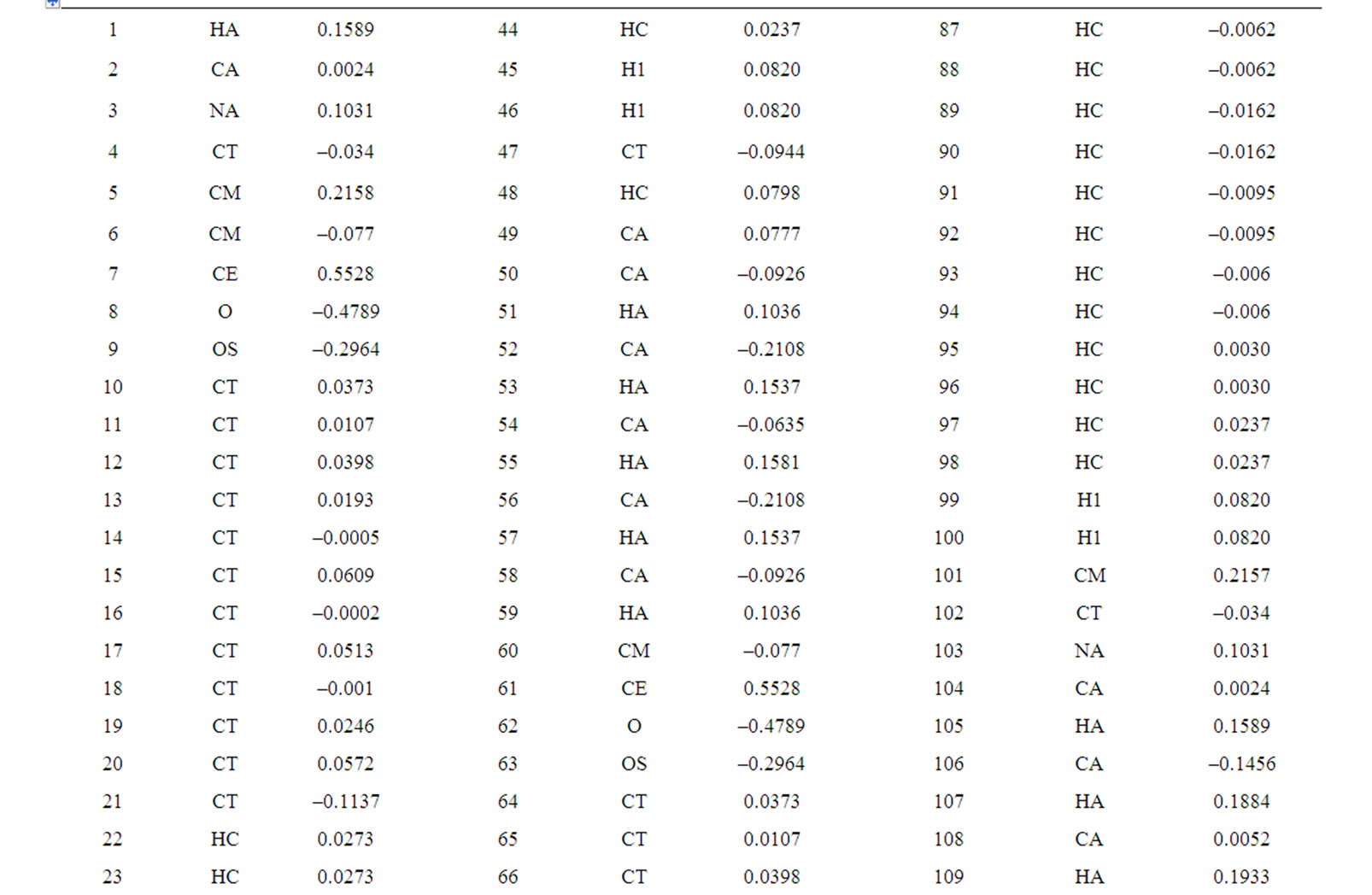

Table S1. Atom charges of DHP-lipid.
Table S2. Derived AMBER parameters for 1,4-DHP lipid.
BOND LENGTH
NA-CT 337.0 1.475
CM-NA 418.0 1.388
CM-CE 410.0 1.444
TORSION ANGLE
CA-NA-CT 70.0 120.00
CA-NA-CA 70.0 120.00
CM-NA-CM 70.0 120.00
CM-NA-H 30.0 118.00
NA-CA-CA 63.0 120.00
NA-CT-CM 63.0 114.00
NA-CT-HC 50.0 109.50
NA-CA-HA 35.0 120.00
CT-CM-NA 70.0 119.70
CM-CM-CE 63.0 120.70
CM-CM-NA 70.0 121.20
CE-CM-CT 70.0 119.70
CM-CE-O 80.0 125.30
CM-CE-OS 80.0 125.30
CM-CT-CA 63.0 114.00
CM-CT-CM 63.0 114.00
DIHEDRAL ANGLE
X-NA-CT-X 6 0.00 0.0 2.0
X-CM-NA-X 4 6.00 180.0 2.0
X-CM-CE-X 4 0.00 0.0 2.0
IMPROPER TORSIONS
CA-CA-NA-CT 1.1 180. 2.0
CT-NA-CM-CM 1.1 180. 2.0
CE-CM-CM-CT 1.1 180. 2.0
CE-CT-CM-CM 1.1 180. 2.0
CM-NA-CM-CT 1.1 180. 2.0
CT-CA-NA-CA 1.1 180. 2.0

