 Journal of Cancer Therapy, 2012, 3, 379-387 http://dx.doi.org/10.4236/jct.2012.324050 Published Online September 2012 (http://www.SciRP.org/journal/jct) 379 Involvement of miR-214 and miR-375 in Malignant Features of Non-Small-Cell Lung Cancer by Down-Regulating CADM1 Megumi Ishimura1, Mika Sakurai-Yageta1, Tomoko Maruyama1, Tomoko Ando2, Masashi Fukayama3, Akiteru Goto1,4, Yoshinori Murakami1* 1Division of Molecular Pathology, The University of Tokyo, Tokyo, Japan; 2Laboratory of Pathology, Institute of Medical Science, The University of Tokyo, Tokyo, Japan; 3Department of Pathology, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan; 4Department of Cellular and Organ Pathology, Graduate School of Medicine, Akita University, Akita, Japan. Email: *ymurakam@ims.u-tokyo.ac.jp Received July 31st, 2012; revised August 30th, 2012; accepted September 13th, 2012 ABSTRACT A tumor suppressor gene, CADM1, encoding an immunoglobulin superfamily cell adhesion molecule, is inactivated in various cancers, including non-small-cell lung cancer (NSCLC). Although promoter methylation is one of the mecha- nisms to suppress CADM1 expression, about half of tumors lacking CADM1 expression do not show methylation of the gene promoter. We herein investigated the possible involvement of microRNA (miRNA) in the down-regulation of CADM1. Using computational algorithms, miR-214 and miR-375 were identified as candidate miRNAs targeting CADM1. A luciferase reporter assay demonstrated that miR-214 and miR-375 repressed the promoter activity through 3’-UTR of CADM1. Quantitative RT-PCR analysis demonstrated that miR-214 and miR-375 was highly expressed in 21 (62%) and 17 (50%) cases of 34 primary NSCLCs. Notably, increased expression of miR-214 was preferentially observed in tumors with advanced pathological stages and in those lacking CADM1 expression but were not associated with the promoter methylation, suggesting that miR-214-mediated silencing would be another mechanism to suppress CADM1 expression. On the other hand, introduction of miR-214 or miR-375 into NSCLC cells decreased CADM1 pro- tein expression. Furthermore, overexpression of miR-214 enhanced anchorage-independent growth of NSCLC cells, A549, whereas transfection of miRNA inhibitors of miR-214 or miR-375 significantly suppressed the in vitro wound healing activity of HCC827 cells. These findings suggest that overexpression of miR-214 and miR-375 could partici- pate in the malignant features of NSCLC through down-regulating CADM1 and would provide a potential target for the treatment of a subset of NSCLC. Keywords: CADM1; miR-214; miR-375; Non-Small-Cell Lung Cancer 1. Introduction Lung cancer is the leading cause of cancer death in the world. More than 80% of lung cancer is categorized as non-small-cell lung cancer (NSCLC). We previously identified a tumor suppressor gene, CADM1/TSLC1, in NSCLC by functional cloning [1]. CADM1 encodes an immunoglobulin superfamily cell-cell adhesion molecule and is expressed in various tissues, including the brain, testis, lung, kidney, and breast [2]. CADM1 expression was frequently lost or reduced in concordance with tumor progression in NSCLC and various other cancers [3]. Moreover, loss or reduction of CADM1 expression indicated worse clinical prognosis of the cases in lung adenocarcinoma, hepatocellular carcinoma, and esopha- geal squamous cell carcinoma [4-7]. However, the me- chanisms underlying the loss of CADM1 expression in cancers are not fully clarified yet. So far, a major postu- lated mechanism is promoter methylation of the CADM1 gene through bi-allelic methylation or mono-allelic me- thylation associated with loss of the other allele. In fact, 44% of NSCLC as well as 30% - 60% of various other cancers showed promoter methylation of the CADM1 gene [8]. However, considerable numbers of tumors lac- king CADM1 expression do not show promoter methy- lation of the CADM1, suggesting that additional un- known mechanisms would be involved in the suppression of CADM1 expression. MicroRNA (miRNA) is a group of small non-coding RNAs that suppress gene expression by interacting with the 3’-UTR of target mRNAs for translational supp- *Corresponding author. Copyright © 2012 SciRes. JCT  Involvement of miR-214 and miR-375 in Malignant Features of Non-Small-Cell Lung Cancer by Down-Regulating CADM1 380 ression and mRNA degradation [9]. Numbers of mi- RNAs were identified to regulate cell proliferation, diffe- rentiation, and apoptosis by targeting different gene sets, thereby acting in an either oncogenic or tumor-supp- ressor manner. Several oncogenic and tumor-suppressor miRNAs have been implicated in the regulation of tumor progression in NSCLC, such as miR-21 and let-7 [10-12]. Therefore, it is tempting to speculate that suppression of CADM1 expression could be mediated by miRNAs in NSCLC. In the present study, we identified miRNAs, miR-214 and miR-375, to target 3’-UTR of CADM1 and examined their functional roles in anchorage-independent cell growth and tumor cell migration/invasion in NSCLC cell lines. Furthermore, we analyzed the expression of miR- 214 and miR-375 in primary NSCLC. Enhancement of the malignant phenotype of NSCLC by these miRNAs, as well as high incidence of their overexpression in pri- mary NSCLC, suggests that miR-214 and miR-375 act as oncomiRs of NSCLC by suppressing CADM1 expres- sion. 2. Materials and Methods 2.1. Cell Lines, Transfection and Clinical Samples A549 and Caco-2 cells were obtained from RIKEN Cell Bank (Ibaraki, Japan); HEK293 from the Health Science Research Resources Bank (Osaka, Japan); and NCI-H441 and HCC827 from The American Type Culture Collec- tion (Manassas, VA). Cells were cultured according to the supplier’s recommendation. Control siRNA (ON- TARGETplus siCONTROL Non-targeting pool, D- 001810-10), siRNA against CADM1 (siGENOME ON- TARGETplus Human IGSF4, J-016565-05), microRNA mimics (miRIDIAN Mimic Human hsa-miR-214, C- 300569-07; miRIDIAN Mimic Human hsa-miR-375, C- 300682-05) and microRNA inhibitors (miRIDIAN Hair- pin Inhibitor Human hsa-miR-214, IH-300569-08; mi- RIDIAN Hairpin Inhibitor Human hsa-miR-375, IH- 300682-07) were obtained from Thermo Fisher Scientific (Waltham, MA). Cells were transfected with 50nM of siRNA or miRNA using LipofectamineTM LTX reagent (Invitrogen). We collected 34 cancer tissues and adjacent non-cancerous lung tissues from NSCLC patients who underwent surgical resection at the University of Tokyo Hospital (Tokyo, Japan) after receiving approval from the Institutional Ethics Review Committee and obtaining informed consent from all patients. 2.2. Real-Time Quantitative PCR (qRT-PCR) Oligonucleotide primer sequences for real-time PCR are shown in Table 1. Total cellular RNA was extracted us- ing an RNeasy Mini kit (QIAGEN Sciences, German- town, MD) and first-strand cDNA was synthesized using a Transcriptor first-strand cDNA synthesis kit (Roche Diagnostics, Basel, Switzerland). Then, real-time PCR was carried out using Light Cycler® 1.5 with light cycler Taq Man master and universal probe of #21 for miRNAs (Roche Diagnostics). Expression of miRNAs was nor- malized to that of small nuclear RNA U6. 2.3. Immunoblot Analysis Immunoblotting was carried out as described previously [2] using a rabbit anti-CADM1 polyclonal antibody (pAb) (C-18) [13] or a goat anti-GAPDH pAb (V-18, Santa Cruz Biotechnology, Santa Cruz, CA) as a control. Quan- tification of signal intensities was performed using Image J Software Ver. 1.44. 2.4. Luciferase Assay CADM1 3’-UTR of 2,848 bp was cloned by RT-PCR Table 1. Primers for cloning of CADM1 3’-UTR and real-time PCR of miRNAs. Target Primer sequence (5’ → 3’) PCR product size (bp) F 5’-GCTAGCATCAGCCTTTTTGTTTCAATGAGG-3’ CADM1 3’-UTR R 5’-ACTAGTCACTTTGTAACATTAATTTTTTTTTATTAAG -3’ 2848 RT 5’-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACACTGCC-3’ miR-214 F 5’-CGGCGGACAGCAGGCACAGACA-3’ 62 RT 5’-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACTCACGC-3’ miR-375 F 5’-CGGCGGTTTGTTCGTTCGGCTC-3’ 62 miR Universal R 5’-GTGCAGGGTCCGAGGT-3’ RT 5’-CGCTTCACGAATTTGCGTGTCAT-3’ F 5’-GTGCTCGCTTCGGCAGCACATATAC-3’ U6 R 5’-CCTTGCGCAGGGGCCATGCTAA-3’ 72 RT: primer for reverse transcriptase reaction; F: forward primer for PCR; R: reverse primer for PCR. Copyright © 2012 SciRes. JCT  Involvement of miR-214 and miR-375 in Malignant Features of Non-Small-Cell Lung Cancer by Down-Regulating CADM1 381 from RNA of Caco-2 cells using primers described in Table 1. The amplified fragment was inserted down- stream of the Firefly luciferase gene of pGL3-Basic (Promega, Madison, WI). Then, a CMV promoter cleaved from pcDNA3.1/Hygro(+) (Invitrogen) was inserted up- stream of the luciferase gene to obtain pCMV-GL3- CADM1-3’-UTR. HEK293 cells were transfected with 25 nM of microRNA mimic, 0.25 μg of pCMV-GL3- CADM1-3’-UTR, and 2.8 ng of pRL-TK (Promega). The luciferase activities of Firefly and Renilla were measured after 48 h with a dual luciferase reporter assay system (Promega) using a Lumat LB9507 luminometer (Ber- thold Technologies, Bad Wildbad, Germany). 2.5. Colony Formation Assay in Soft Agar After 48 h of transfection, A549 cells were trypsinized and plated at 1 × 104 in 0.36% soft agar on the top of a base layer containing 0.5% agar and grown at 37˚C for 15 days. The numbers of colonies per dish with diame- ters over 100 m formed in soft agar were counted under the microscope and normalized to those in a liquid cul- ture for 7 days. 2.6. Wound Healing Assay HCC827 cells transfected with 50 nM of microRNA in- hibitors or 1 μg of the CADM1 expression vector were cultured to obtain 90% confluency. A wound was created by scraping the cells using a P200 pipette tip and images were captured immediately (0 h) and at 10 h and 20 h. Migration of cells was assessed by measuring the aver- age width of the wounds at three different points. 2.7. Immunohistochemical Analysis Immunohistochemistry was performed using anti-CADM1 polyclonal antibody, CC2, as described previously [5]. 2.8. Methylation-Specific PCR (MSP) Methylation-specific PCR of the CADM1 promoter was performed as described elsewhere [14]. The primers used were: 5’-AGTGACGGAAATTTGTAACG-3’ and 5’- AAAAACTCGAACTCCAAAAAACG-3’ for the me- thylated DNA and 5’-AGTGATGGAAATTTGTAATG- 3’ and 5’-AAAAACTCAAACTCCAAAAAACA-3’ for unmethylated DNA. 2.9. Statistical Analysis Statistical differences were determined by the two-tailed Student’s t-test with SigmaPlot software Ver. 11 (Systat Software, San Jose, CA). Fisher’s exact tests were app- lied to test significant associations between the expres- sion state of miRNAs and the clinicopathological char- acteristics of primary NSCLC. A p-value of <0.05 was considered as statistically significant. 3. Results 3.1. Identification of miRNAs Targeting CADM1 A couple of computational algorithms TargetScan [15] and PicTar [16], were used to identify a numbers of po- tential miRNAs targeting CADM1. The search program TargetScan identified miR-375 and miR-214 (Table 2) as the strongest candidate miRNA targeting CADM1, while another program, PicTar, identified miR-214 (Ta- ble 3). Database analysis showed that three and two pre- dicted binding sites of miR-214 and miR-375 were pre- sent, respectively, in the 3’-UTR of the CADM1 (Figure 1(a)). The seed sequences (SS) of miR-214 and miR-375 were well conserved between mouse and human, except for the third predicted seed sequences of miR-214-SS3 located about the 1200 bp from the starting site of the CADM1 3’-UTR (Figure 1(b)). Table 2. Prediction of miRNAs targeting CADM1 by Tar- getScan Human 5.2. miRNA Aggregate PCT a miR-375 0.50 miR-214/761 0.42 miR-124/506 0.37 miR-129/129-5p 0.33 miR-205 0.30 miR-148/152 0.28 miR-101 0.27 miR-138 0.26 miR-200bc/429 0.22 miR-208/208ab 0.19 aPCT, Preferentially conserved targeting. Table 3. Prediction of miRNAs targeting CADM1 by Pic- Tar. miRNA PicTar Score miR-214 10.25 miR-182* 8.57 miR-144 3.94 miR-199a* 3.94 miR-101 3.60 miR-190 2.47 miR-186 2.47 miR-10b 2.29 miR-10a 2.09 miR-195 1.87 Copyright © 2012 SciRes. JCT 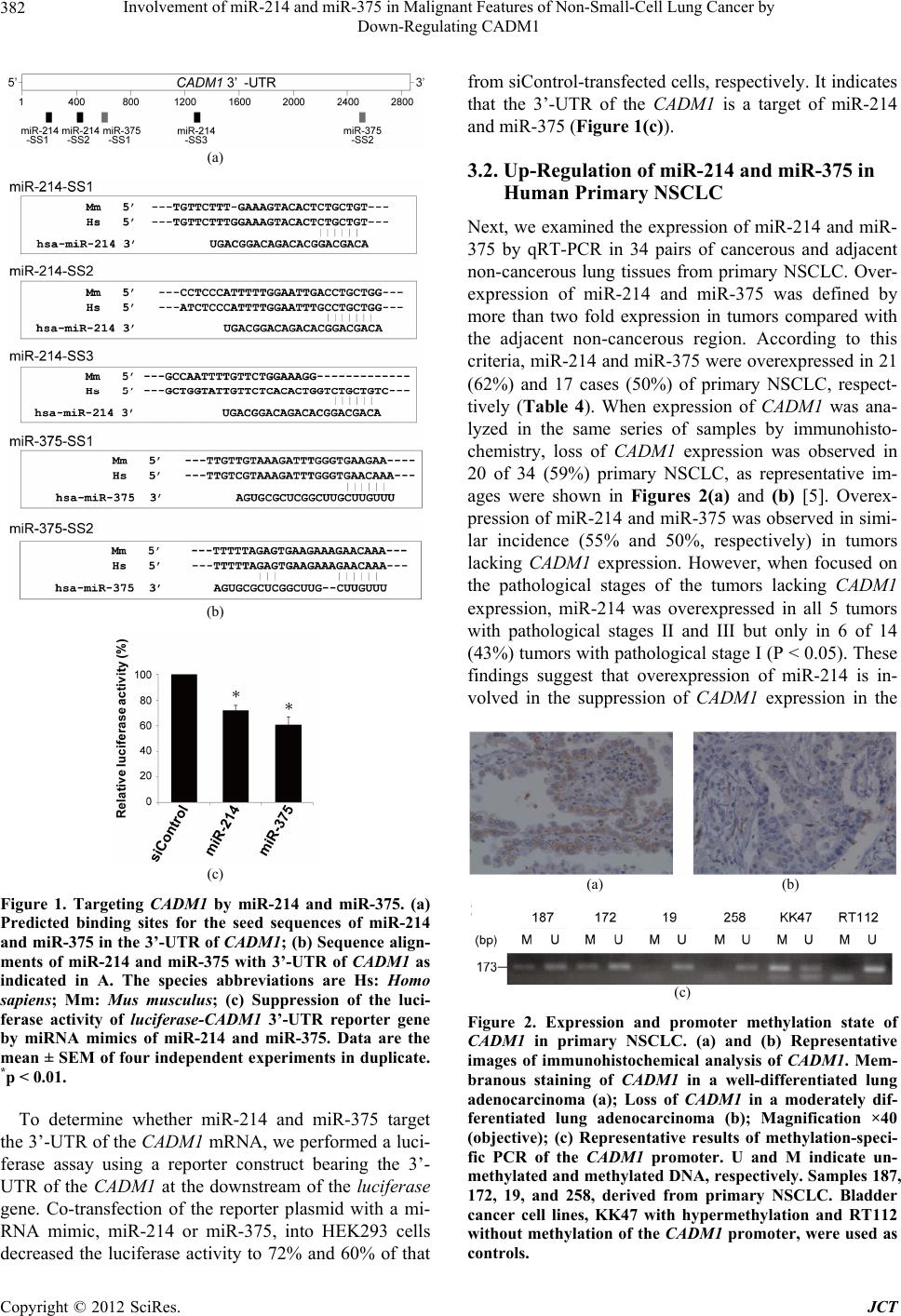 Involvement of miR-214 and miR-375 in Malignant Features of Non-Small-Cell Lung Cancer by Down-Regulating CADM1 382 (a) (b) (c) Figure 1. Targeting CADM1 by miR-214 and miR-375. (a) Predicted binding sites for the seed sequences of miR-214 and miR-375 in the 3’-UTR of CADM1; (b) Sequence align- ments of miR-214 and miR-375 with 3’-UTR of CADM1 as indicated in A. The species abbreviations are Hs: Homo sapiens; Mm: Mus musculus; (c) Suppression of the luci- ferase activity of luciferase-CADM1 3’-UTR reporter gene by miRNA mimics of miR-214 and miR-375. Data are the mean ± SEM of four independent experiments in duplicate. *p < 0.01. To determine whether miR-214 and miR-375 target the 3’-UTR of the CADM1 mRNA, we performed a luci- ferase assay using a reporter construct bearing the 3’- UTR of the CADM1 at the downstream of the luciferase gene. Co-transfection of the reporter plasmid with a mi- RNA mimic, miR-214 or miR-375, into HEK293 cells decreased the luciferase activity to 72% and 60% of that from siControl-transfected cells, respectively. It indicates that the 3’-UTR of the CADM1 is a target of miR-214 and miR-375 (Figure 1(c)). 3.2. Up-Regulation of miR-214 and miR-375 in Human Primary NSCLC Next, we examined the expression of miR-214 and miR- 375 by qRT-PCR in 34 pairs of cancerous and adjacent non-cancerous lung tissues from primary NSCLC. Over- expression of miR-214 and miR-375 was defined by more than two fold expression in tumors compared with the adjacent non-cancerous region. According to this criteria, miR-214 and miR-375 were overexpressed in 21 (62%) and 17 cases (50%) of primary NSCLC, respect- tively (Table 4). When expression of CADM1 was ana- lyzed in the same series of samples by immunohisto- chemistry, loss of CADM1 expression was observed in 20 of 34 (59%) primary NSCLC, as representative im- ages were shown in Figures 2(a) and (b) [5]. Overex- pression of miR-214 and miR-375 was observed in simi- lar incidence (55% and 50%, respectively) in tumors lacking CADM1 expression. However, when focused on the pathological stages of the tumors lacking CADM1 expression, miR-214 was overexpressed in all 5 tumors with pathological stages II and III but only in 6 of 14 (43%) tumors with pathological stage I (P < 0.05). These findings suggest that overexpression of miR-214 is in- volved in the suppression of CADM1 expression in the (a) (b) (c) Figure 2. Expression and promoter methylation state of CADM1 in primary NSCLC. (a) and (b) Representative images of immunohistochemical analysis of CADM1. Mem- branous staining of CADM1 in a well-differentiated lung adenocarcinoma (a); Loss of CADM1 in a moderately dif- ferentiated lung adenocarcinoma (b); Magnification ×40 (objective); (c) Representative results of methylation-speci- fic PCR of the CADM1 promoter. U and M indicate un- methylated and methylated DNA, respectively. Samples 187, 172, 19, and 258, derived from primary NSCLC. Bladder cancer cell lines, KK47 with hypermethylation and RT112 without methylation of the CADM1 promoter, were used as controls. Copyright © 2012 SciRes. JCT 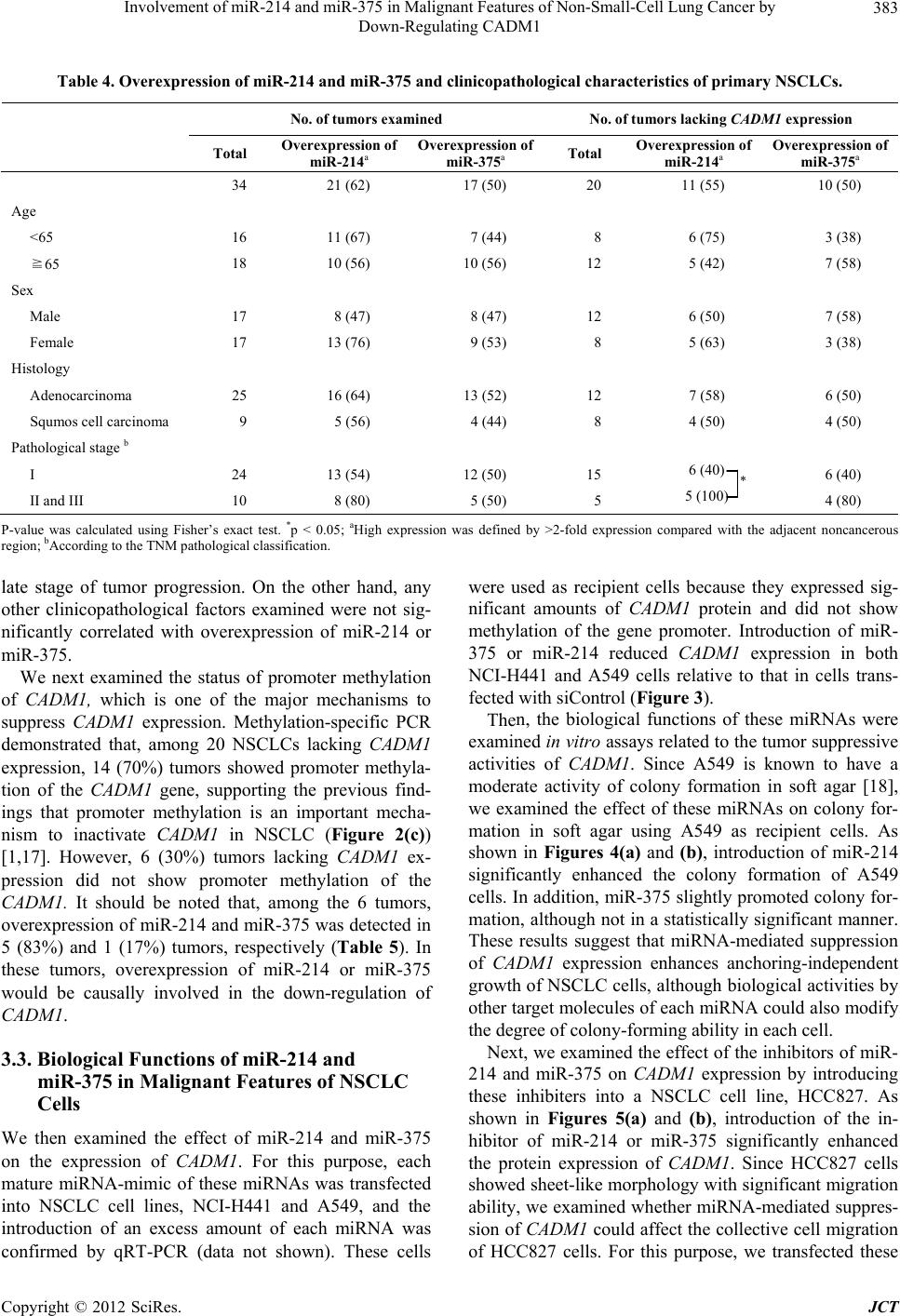 Involvement of miR-214 and miR-375 in Malignant Features of Non-Small-Cell Lung Cancer by Down-Regulating CADM1 Copyright © 2012 SciRes. JCT 383 Table 4. Overexpression of miR-214 and miR-375 and clinicopathological characteristics of primary NSCLCs. No. of tumors examined No. of tumors lacking CADM1 expression Total Overexpression of miR-214a Overexpression of miR-375a Total Overexpression of miR-214a Overexpression of miR-375a 34 21 (62) 17 (50) 20 11 (55) 10 (50) Age <65 16 11 (67) 7 (44) 8 6 (75) 3 (38) ≧65 18 10 (56) 10 (56) 12 5 (42) 7 (58) Sex Male 17 8 (47) 8 (47) 12 6 (50) 7 (58) Female 17 13 (76) 9 (53) 8 5 (63) 3 (38) Histology Adenocarcinoma 25 16 (64) 13 (52) 12 7 (58) 6 (50) Squmos cell carcinoma 9 5 (56) 4 (44) 8 4 (50) 4 (50) Pathological stage b I 24 13 (54) 12 (50) 15 6 (40) 6 (40) II and III 10 8 (80) 5 (50) 5 5 (100) 4 (80) * P-value was calculated using Fisher’s exact test. *p < 0.05; aHigh expression was defined by >2-fold expression compared with the adjacent noncancerous region; bAccording to the TNM pathological classification. were used as recipient cells because they expressed sig- nificant amounts of CADM1 protein and did not show methylation of the gene promoter. Introduction of miR- 375 or miR-214 reduced CADM1 expression in both NCI-H441 and A549 cells relative to that in cells trans- fected with siControl (Figure 3). late stage of tumor progression. On the other hand, any other clinicopathological factors examined were not sig- nificantly correlated with overexpression of miR-214 or miR-375. We next examined the status of promoter methylation of CADM1, which is one of the major mechanisms to suppress CADM1 expression. Methylation-specific PCR demonstrated that, among 20 NSCLCs lacking CADM1 expression, 14 (70%) tumors showed promoter methyla- tion of the CADM1 gene, supporting the previous find- ings that promoter methylation is an important mecha- nism to inactivate CADM1 in NSCLC (Figure 2(c)) [1,17]. However, 6 (30%) tumors lacking CADM1 ex- pression did not show promoter methylation of the CADM1. It should be noted that, among the 6 tumors, overexpression of miR-214 and miR-375 was detected in 5 (83%) and 1 (17%) tumors, respectively (Table 5). In these tumors, overexpression of miR-214 or miR-375 would be causally involved in the down-regulation of CADM1. Then, the biological functions of these miRNAs were examined in vitro assays related to the tumor suppressive activities of CADM 1. Since A549 is known to have a moderate activity of colony formation in soft agar [18], we examined the effect of these miRNAs on colony for- mation in soft agar using A549 as recipient cells. As shown in Figures 4(a) and (b), introduction of miR-214 significantly enhanced the colony formation of A549 cells. In addition, miR-375 slightly promoted colony for- mation, although not in a statistically significant manner. These results suggest that miRNA-mediated suppression of CAD M1 expression enhances anchoring-independent growth of NSCLC cells, although biological activities by other target molecules of each miRNA could also modify the degree of colony-forming ability in each cell. Next, we examined the effect of the inhibitors of miR- 214 and miR-375 on CADM1 expression by introducing these inhibiters into a NSCLC cell line, HCC827. As shown in Figures 5(a) and (b), introduction of the in- hibitor of miR-214 or miR-375 significantly enhanced the protein expression of CADM1 . Since HCC827 cells showed sheet-like morphology with significant migration ability, we examined whether miRNA-mediated suppres- sion of CADM1 could affect the collective cell migration of HCC827 cells. For this purpose, we transfected these 3.3. Biological Functions of miR-214 and miR-375 in Malignant Features of NSCLC Cells We then examined the effect of miR-214 and miR-375 on the expression of CADM1. For this purpose, each mature miRNA-mimic of these miRNAs was transfected into NSCLC cell lines, NCI-H441 and A549, and the introduction of an excess amount of each miRNA was confirmed by qRT-PCR (data not shown). These cells  Involvement of miR-214 and miR-375 in Malignant Features of Non-Small-Cell Lung Cancer by Down-Regulating CADM1 384 Table 5. Overexpression of miR-214 and miR-375 and expression and promoter methylation state of CADM1 in primary NSCLCs. No. of tumors examined No. of tumors lacking CADM1 expression Total Overexpression of miR-214 a Overexpression of miR-375 a Total Overexpression of miR-214 a Overexpression of miR-375 a Methylation state Methylated 17 9 (53) 10 (59) 14 6 (43) 9 (64) Unmethylated 17 12 (71) 7 (41) 6 5 (83) 1 (17) a High expression was defined by >2-fold expression compared with the adjacent noncancerous region. (a) (b) Figure 3. Effects of miRNA mimics and inhibitors of miR- 214 and miR-375 on CADM1 expression in NSCLC cells. Immunoblotting (a) and its quantification (b) of NCI-H441 (left) and A549 cells (right) transfected with siControl, si- CADM1, or miRNA mimics as indicated; (b) Intensities of CADM1 were normalized to those of GAPDH in NCI-H441 (black) and A549 (white) cells. Relative intensities to siCon- trol-transfected cells are shown. Data are the mean ± SD of four independent experiments. **p < 0.05; *p < 0.01. inhibitors of miRNAs as well as an expression vector of CADM1 into HCC827 cells and assessed their wound- healing activity. As shown in Figures 5(c) and (d), the velocity of HCC827 cells transfected with inhibitors of miR-214 and miR-375 significantly decreased to 64% and 66% of that of siControl-transfected cells, respect- tively, with values that are almost equivalent to those seen in HCC827 transfected with CADM1 (50%). These results suggest that CADM1 is one of target genes of miR-214 and miR-375 for their enhancement in collec- tive cell migration of an NSCLC cell, HCC827. 4. Discussion In this study, using a reporter assay, suppression of CADM1 expression by miR-214 was demonstrated for (a) (b) Figure 4. Soft agar colony formation of A549 cells trans- fected with miRNA mimics of miR-214 or miR-375. (a) Representative images of A549 cells with miRNA mimics as indicated. Bars, 200 m; (b) Numbers of relevant colonies are shown relative to those of control cells. Data are the mean ± SD of three independent experiments. **p < 0.05. the first time. Moreover, miR-375-mediated CADM1 suppression is consistent with the previous report that CADM1 is one of the 10 targets of miR-375 in pancre- atic islet cells derived from miR-375-deficient mice [19]. It is noteworthy that CADM1 gives a pair of transcript with the molecular size of 4.4 kb and 1.6 kb by Northern blot analysis [1], but that the shorter transcript does not contain the sequences in its 3’-UTR targeted by miR-214 or miR-375. In normal lung, as well as many other tis- sues, the transcript of 4.4 kb is predominantly expressed and could be physiologically regulated by these miRNAs. On the other hand, the ratio of 4.4 kb-transcript to 1.6 kb-transcript was significantly decreased in several NSCLC cells in comparison with normal lung tissues, suggesting that CADM1 transcript of 4.4 kb is selectively degraded by overexpressed miR-214 and miR-375 in NSCLC cells. Copyright © 2012 SciRes. JCT 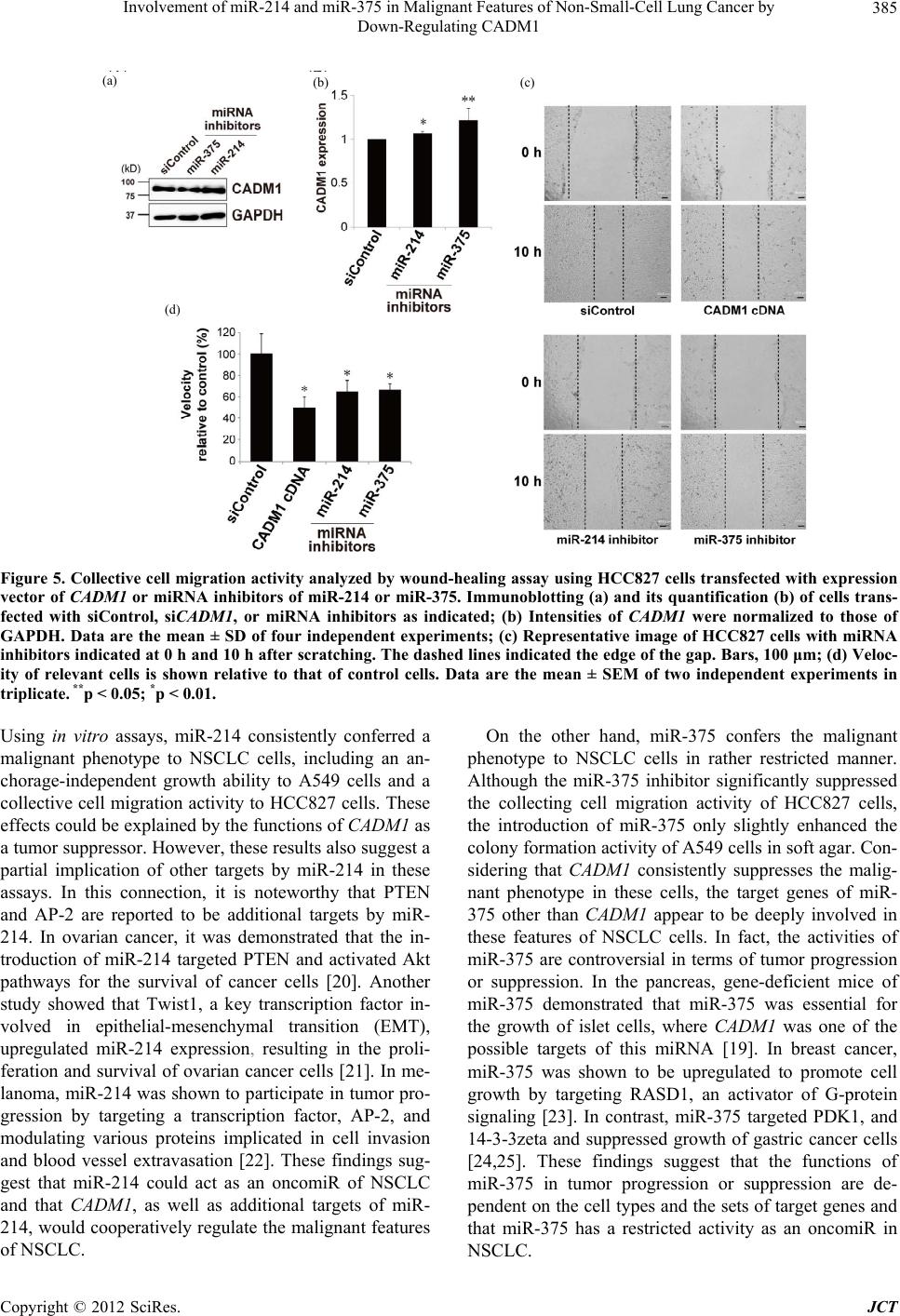 Involvement of miR-214 and miR-375 in Malignant Features of Non-Small-Cell Lung Cancer by Down-Regulating CADM1 385 (a) (b) (c) (d) Figure 5. Collective cell migration activity analyzed by wound-healing assay using HCC827 cells transfected with expression vector of CADM1 or miRNA inhibitors of miR-214 or miR-375. Immunoblotting (a) and its quantification (b) of cells trans- fected with siControl, siCADM1, or miRNA inhibitors as indicated; (b) Intensities of CADM1 were normalized to those of GAPDH. Data are the mean ± SD of four independent experiments; (c) Representative image of HCC827 cells with miRNA inhibitors indicated at 0 h and 10 h after scratching. The dashed lines indicated the edge of the gap. Bars, 100 μm; (d) Veloc- ity of relevant cells is shown relative to that of control cells. Data are the mean ± SEM of two independent experiments in triplicate. **p < 0.05; *p < 0.01. Using in vitro assays, miR-214 consistently conferred a malignant phenotype to NSCLC cells, including an an- chorage-independent growth ability to A549 cells and a collective cell migration activity to HCC827 cells. These effects could be explained by the functions of CADM1 as a tumor suppressor. However, these results also suggest a partial implication of other targets by miR-214 in these assays. In this connection, it is noteworthy that PTEN and AP-2 are reported to be additional targets by miR- 214. In ovarian cancer, it was demonstrated that the in- troduction of miR-214 targeted PTEN and activated Akt pathways for the survival of cancer cells [20]. Another study showed that Twist1, a key transcription factor in- volved in epithelial-mesenchymal transition (EMT), upregulated miR-214 expression, resulting in the proli- feration and survival of ovarian cancer cells [21]. In me- lanoma, miR-214 was shown to participate in tumor pro- gression by targeting a transcription factor, AP-2, and modulating various proteins implicated in cell invasion and blood vessel extravasation [22]. These findings sug- gest that miR-214 could act as an oncomiR of NSCLC and that CADM1, as well as additional targets of miR- 214, would cooperatively regulate the malignant features of NSCLC. On the other hand, miR-375 confers the malignant phenotype to NSCLC cells in rather restricted manner. Although the miR-375 inhibitor significantly suppressed the collecting cell migration activity of HCC827 cells, the introduction of miR-375 only slightly enhanced the colony formation activity of A549 cells in soft agar. Con- sidering that CADM1 consistently suppresses the malig- nant phenotype in these cells, the target genes of miR- 375 other than CADM1 appear to be deeply involved in these features of NSCLC cells. In fact, the activities of miR-375 are controversial in terms of tumor progression or suppression. In the pancreas, gene-deficient mice of miR-375 demonstrated that miR-375 was essential for the growth of islet cells, where CADM1 was one of the possible targets of this miRNA [19]. In breast cancer, miR-375 was shown to be upregulated to promote cell growth by targeting RASD1, an activator of G-protein signaling [23]. In contrast, miR-375 targeted PDK1, and 14-3-3zeta and suppressed growth of gastric cancer cells [24,25]. These findings suggest that the functions of miR-375 in tumor progression or suppression are de- pendent on the cell types and the sets of target genes and that miR-375 has a restricted activity as an oncomiR in NSCLC. Copyright © 2012 SciRes. JCT  Involvement of miR-214 and miR-375 in Malignant Features of Non-Small-Cell Lung Cancer by Down-Regulating CADM1 386 In addition to the cell biological analyses, we exam- ined the expression of miR-214 and miR-375 in 34 pri- mary NSCLCs in this study. As discussed above, it is necessary to consider the patho-biological significance of these miRNAs from two viewpoints: one, as an inde- pendent oncomiR with multiple distinct target molecules, and the other, as miRNA that targets a tumor suppressor, CADM1. The high incidences of overexpression of miR- 214 (62%) and miR-375 (50%) in primary NSCLC strongly suggest that both miR-214 and miR-375 act as oncomiRs in human NSCLC. MiR-214 was upregulated in primary ovarian cancer [20], whereas higher expres- sion of miR-375 was associated with poor prognosis of esophageal adenocarcinoma [26]. MiR-375 was also re- ported as a candidate sputum biomarker of lung adeno- carcinoma [27]. On the other hand, with regard to the ability of mi- RNAs to target a tumor suppressor, CADM1, overex- pression of these miRNAs could provide a novel mo- lecular mechanism to suppress CADM1 expression in NSCLC in addition to the methylation of the gene pro- moter. In fact, among the tumors lacking CADM1 ex- pression, miR-214 overexpression is observed in much higher incidence in tumors without the CADM1 methyla- tion in comparison with tumors with the CADM1 methy- lation. Furthermore, the high incidence of overexpression of miR-214 in advanced NSCLC with pathological stages II and III (P < 0.05). It suggests that disruption of miRNA expression could be a rather late event in multi- stage tumorigenesis of NSCLC. 5. Conclusion We demonstrate a novel mechanism to suppress the ex- pression of a tumor suppressor CADM1 by miRNAs, miR-214 and miR-375, in NSCLC. Overexpression of miR-214 and miR-375 in more than half of primary NSCLC, as well as the enhancement in cell motility and anchorage-independent cell growth of NSCLC cells by miR-214 or miR-375 through suppression of CADM1 expression, suggests that miR-214 and miR-375 would provide potential targets for the treatment of a subset of NSCLC. 6. Acknowledgements The authors express their gratitude to Dr. Junichiro Inoue, Division of Cellular and Molecular Biology, Department of Cancer Biology, Institute of Medical Science, the University of Tokyo, for his generous help with the Lu- mat LB9507 luminometer. We thank members of Divi- sion of Molecular Pathology for constructive criticism and fruitful discussion. This work was supported in part by a Grant-in-Aid for Young Scientists (B) [24790310 for M.SY.] and a Grant-in-Aid for Scientific Research (B) [22300336 for Y.M.] from the Ministry of Education, Culture, Sports, Science, and Technology, Japan; a Grant-in-Aid for the Third Term Comprehensive Control Research for Cancer from the Ministry of Health, Labor, and Welfare, Japan [Y.M.]; and a Grant from the Smok- ing Research Foundation, Japan [Y.M.]. REFERENCES [1] M. Kuramochi, H. Fukuhara, T. Nobukuni, T. Kanbe, T. Maruyama, H. P. Ghosh, M. Pletcher, M. Isomura, M. Onizuka, T. Kitamura, T. Sekiya, R. H. Reeves and Y. Murakami, “TSLC1 Is a Tumor-Suppressor Gene in Hu- man Non-Small-Cell Lung Cancer,” Nature Genetics, Vol. 27, No. 4, 2001, pp. 427-430. [2] M. Masuda, M. Yageta, H. Fukuhara, M. Kuramochi, T. Maruyama, A. Nomoto and Y. Murakami, “The Tumor Suppressor Protein TSLC1 Is Involved in Cell-Cell Ad- hesion,” Journal of Biological Chemistry, Vol. 277, No. 34, 2002, pp. 31014-31019. [3] T. Fukami, H. Fukuhara, M. Kuramochi, T. Maruyama, K. Isogai, M. Sakamoto, S. Takamoto and Y. Murakami, “Promoter Methylation of the TSLC1 Gene in Advanced Lung Tumors and Various Cancer Cell Lines,” Interna- tional Journal of Cancer, Vol. 107, No. 1, 2003, pp. 53- 59. doi:10.1002/ijc.11348 [4] K. Uchino, A. Ito, T. Wakayama, Y. Koma, T. Okada, C. Ohbayashi, S. Iseki, Y. Kitamura, N. Tsubota, Y. Okita and M. Okada, “Clinical Implication and Prognostic Sig- nificance of the Tumor Suppressor TSLC1 Gene Detected in Adenocarcinoma of the Lung,” Cancer, Vol. 98, No. 5, 2003, pp. 1002-1007. doi:10.1002/cncr.11599 [5] A. Goto, T. Niki, L. Chi-Pin, D. Matsubara, Y. Murakami, N. Funata and M. Fukayama, “Loss of TSLC1 Expression in Lung Adenocarcinoma: Relationships with Histologi- cal Subtypes, Sex and Prognostic Significance,” Cancer Science, Vol. 96, No. 8, 2005, pp. 480-486. doi:10.1111/j.1349-7006.2005.00075.x [6] W. Zhang, L. Zhou, S. M. Ding, H. Y. Xie, X. Xu, J. Wu, Q. X. Chen, F. Zhang, B. J. Wei, A. T. Eldin and S. S. Zheng, “Aberrant Methylation of the CADM1 Promoter Is Associated with Poor Prognosis in Hepatocellular Carci- noma Treated with Liver Transplantation,” Oncology Reports, Vol. 25, No. 4, 2011, pp. 1053-1062. doi:10.3892/or.2011.1159 [7] T. Ito, Y. Shimada, Y. Hashimoto, J. Kaganoi, T. Kan, G. Watanabe, Y. Murakami and M. Imamura, “Involvement of TSLC1 in Progression of Esophageal Squamous Cell Carcinoma,” Cancer Research, Vol. 63, No. 19, 2003, pp. 6320-6326. [8] Y. Murakami, “Involvement of a Cell Adhesion Molecule, TSLC1/IGSF4, in Human Oncogenesis,” Cancer Science, Vol. 96, No. 9, 2005, pp. 543-552. doi: 10.1111/j.1349-7006.2005.00089.x [9] D. P. Bartel, “MicroRNAs: Genomics, Biogenesis, Me- chanism, and Function,” Cell, Vol. 116, No. 2, 2004, pp. 281-297. Copyright © 2012 SciRes. JCT  Involvement of miR-214 and miR-375 in Malignant Features of Non-Small-Cell Lung Cancer by Down-Regulating CADM1 Copyright © 2012 SciRes. JCT 387 [10] M. Seike, A. Goto, T. Okano, E. D. Bowman, A. J. Schetter, I. Horikawa, E. A. Mathe, J. Jen, P. Yang, H. Sugimura, A. Gemma, S. Kudoh, C. M. Croce and C. C. Harris, “MiR-21 Is an EGFR-Regulated Anti-Apoptotic Factor in Lung Cancer in Never-Smokers,” Proceedings of the National Academy of Science of the United States of America, Vol. 106, No. 29, 2009, pp. 12085-12090. doi:10.1073/pnas.0905234106 [11] F. Talotta, A. Cimmino, M. R. Matarazzo, L. Casalino, G. De Vita, M. D’Esposito, R. Di Lauro and P. Verde, “An Autoregulatory Loop Mediated by miR-21 and PDCD4 Controls the AP-1 Activity in RAS Transformation,” Oncogene, Vol. 28, No. 1, 2009, pp. 73-84. doi:10.1038/onc.2008.370 [12] S. M. Johnson, H. Grosshans, J. Shingara, M. Byrom, R. Jarvis, A. Cheng, E. Labourier, K. L. Reinert, D. Brown and F. J. Slack, “RAS Is Regulated by the Let-7 micro- RNA Family,” Cell, Vol. 120, No. 5, 2005, pp. 635-647. doi:10.1016/j.cell.2005.01.014 [13] T. Ito, Y. Williams-Nate, M. Iwai, Y. Tsuboi, M. Hagi- yama, A. Ito, M. Sakurai-Yageta and Y. Murakami, “Transcriptional Regulation of the CADM1 Gene by Retinoic Acid during the Neural Differentiation of Mur- ine Embryonal Carcinoma P19 Cells,” Genes to Cells, Vol. 16, No. 7, 2011, pp. 791-802. doi:10.1111/j.1365-2443.2011.01525.x [14] J. G. Herman, J. R. Graff, S. Myohanen, B. D. Nelkin and S. B. Baylin, “Methylation-Specific PCR: A Novel PCR Assay for Methylation Status of CpG Islands,” Proceed- ings of the National Academy of Science of the United States of America, Vol. 93, No. 18, 1996, pp. 9821-9826. [15] B. P. Lewis, I. H. Shih, M. W. Jones-Rhoades, D. P. Bartel and C. B. Burge, “Prediction of Mammalian mi- croRNA Targets,” Cell, Vol. 115, No. 7, 2003, pp. 787- 798. [16] A. Krek, D. Grun, M. N. Poy, R. Wolf, L. Rosenberg, E. J. Epstein, P. MacMenamin, I. da Piedade, K. C. Gunsalus, M. Stoffel and N. Rajewsky, “Combinatorial microRNA Target Predictions,” Nature Genetics, Vol. 37, No. 5, 2005, pp. 495-500. doi:10.1038/ng1536 [17] S. Kikuchi, D. Yamada, T. Fukami, T. Maruyama, A. Ito, H. Asamura, Y. Matsuno, M. Onizuka and Y. Murakami. “Hypermethylation of the TSLC1/IGSF4 Promoter Is Associated with Tobacco Smoking and a Poor Prognosis in Primary Non-Small Cell Lung Cancer,” Cancer, Vol. 106, No. 8, 2006, pp. 1751-1758. doi:10.1002/cncr.21800 [18] X. Mao, E. Seidlitz, K. Ghosh, Y. Murakami and H. P. Ghosh, “The Cytoplasmic Domain Is Critical to the Tu- mor Suppressor Activity of TSLC1 in Non-Small Cell Lung Cancer,” Cancer Research, Vol. 63, No. 22, 2003, pp. 7979-7985. [19] M. N. Poy, J. Hausser, M. Trajkovski, M. Braun, S. Collins, P. Rorsman, M. Zavolan and M. Stoffel, “miR- 375 Maintains Normal Pancreatic Alpha- and Beta-Cell Mass,” Proceedings of the National Academy of Science of the United States of America, Vol. 106, No. 14, 2009, pp. 5813-5818. doi:10.1073/pnas.0810550106 [20] H. Yang, W. Kong, L. He, J. J. Zhao, J. D. O’Donnell, J. Wang, R. M. Wenham, D. Coppola, P. A. Kruk, S. V. Nicosia and J. Q. Cheng, “MicroRNA Expression Profil- ing in Human Ovarian Cancer: miR-214 Induces Cell Survival and Cisplatin Resistance by Targeting PTEN,” Cancer Research, Vol. 68, No. 2, 2008, pp. 425-433. doi:10.1158/0008-5472.CAN-07-2488 [21] G. Yin, R. Chen, A. B. Alvero, H. H. Fu, J. Holmberg, C. Glackin, T. Rutherford and G. Mor, “TWISTing Stem- ness, Inflammation and Proliferation of Epithelial Ovar- ian Cancer Cells through MIR199A2/214,” Oncogene, Vol. 29, No. 24, 2010, pp. 3545-3553. doi:10.1038/onc.2010.111 [22] E. Penna, F. Orso, D. Cimino, E. Tenaglia, A. Lembo, E. Quaglino, L. Poliseno, A. Haimovic, S. Osella-Abate, C. De Pitta, E. Pinatel, M. B. Stadler, P. Provero, M. G. Bernengo, I. Osman and D. Taverna, “microRNA-214 Contributes to Melanoma Tumour Progression through Suppression of TFAP2C,” EMBO Journal, Vol. 30, No.10, 2011, pp. 1990-2007. doi:10.1038/emboj.2011.102 [23] P. de Souza Rocha Simonini, A. Breiling, N. Gupta, M. Malekpour, M. Youns, R. Omranipour, F. Malekpour, S. Volinia, C. M. Croce, H. Najmabadi, S. Diederichs, O. Sahin, D. Mayer, F. Lyko, J. D. Hoheisel and Y. Riazal- hosseini, “Epigenetically Deregulated microRNA-375 Is Involved in a Positive Feedback Loop with Estrogen Re- ceptor Alpha in Breast Cancer Cells,” Cancer Research, Vol. 70, No. 22, 2010, pp. 9175-9184. doi:10.1158/0008-5472.CAN-10-1318 [24] Y. Tsukamoto, C. Nakada, T. Noguchi, M. Tanigawa, L. T. Nguyen, T. Uchida, N. Hijiya, K. Matsuura, T. Fujioka, M. Seto and M. Moriyama, “MicroRNA-375 Is Down- regulated in Gastric Carcinomas and Regulates Cell Sur- vival by Targeting PDK1 and 14-3-3zeta,” Cancer Re- search, Vol. 70, No. 6, 2010, pp. 2339-2349. doi:10.1158/0008-5472.CAN-09-2777 [25] L. Ding, Y. Xu, W. Zhang, Y. Deng, M. Si, Y. Du, H. Yao, X. Liu, Y. Ke, J. Si and T. Zhou, “MiR-375 Fre- quently Downregulated in Gastric Cancer Inhibits Cell Proliferation by Targeting JAK2,” Cell Resea rch, Vol. 20, No. 7, 2010, pp. 784-793. doi: 10.1038/cr.2010.79 [26] E. A. Mathe, G. H. Nguyen, E. D. Bowman, Y. Zhao, A. Budhu, A. J. Schetter, R. Braun, M. Reimers, K. Kuma- moto, D. Hughes, N. K. Altorki, A. G. Casson, C. G. Liu, X. W. Wang, N. Yanaihara, N. Hagiwara, A. J. Dannen- berg, M. Miyashita, C. M. Croce and C. C. Harris, “Mi- croRNA Expression in Squamous Cell Carcinoma and Adenocarcinoma of the Esophagus: Associations with Survival,” Clinical Cancer Research, Vol. 15, No. 19, 2009, pp. 6192-6200. doi:10.1158/1078-0432.CCR-09-1467 [27] L. Yu, N. W. Todd, L. Xing, Y. Xie, H. Zhang, Z. Liu, H. Fang, J. Zhang, R. L. Katz and F. Jiang, “Early Detection of Lung Adenocarcinoma in Sputum by a Panel of mi- croRNA Markers,” International Journal of Cancer, Vol. 127, No. 12, 2010, pp. 2870-2878. doi:10.1002/ijc.25289
|