Journal of Electromagnetic Analysis and Applications
Vol.07 No.05(2015), Article ID:56404,15 pages
10.4236/jemaa.2015.75017
A Comparative Study of Automated Segmentation Methods for Use in a Microwave Tomography System for Imaging Intracerebral Hemorrhage in Stroke Patients
Qaiser Mahmood1,2, Shaochuan Li1, Andreas Fhager1,2, Stefan Candefjord1,2, Artur Chodorowski1,2, Andrew Mehnert3, Mikael Persson1,2
1Department of Signals and Systems, Chalmers University of Technology, Gothenburg, Sweden
2MedTech West, Sahlgrenska University Hospital, Gothenburg, Sweden
3Centre for Microscopy, Characterisation and Analysis, The University of Western Australia, Perth, Australia
Email: qaiserm@chalmers.se, stephen.li1126@gmail.com, andreas.fhager@chalmers.se, stefan.candefjord@chalmers.se, atur.chodorowski@chalmers.se, andrew.mehnert@uwa.edu.au, mikael.persson@chalmers.se
Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 30 March 2015; accepted 16 May 2015; published 19 May 2015
ABSTRACT
Microwave technology offers the possibility for pre-hospital stroke detection as we have previously demonstrated using non-imaging diagnostics. The focus in this paper is on image-based diagnostics wherein the technical and computational complexities of image reconstruction are a challenge for clinical realization. Herein we investigate whether information about a patient’s brain anatomy obtained prior to a stroke event can be used to facilitate image-based stroke diagnostics. A priori information can be obtained by segmenting the patient’s head tissues from magnetic resonance images. Expert manual segmentation is presently the gold standard, but it is laborious and subjective. A fully automatic method is thus desirable. This paper presents an evaluation of several such methods using both synthetic magnetic resonance imaging (MRI) data and real data from four healthy subjects. The segmentation was performed on the full 3D MRI data, whereas the electromagnetic evaluation was performed using a 2D slice. The methods were evaluated in terms of: i) tissue classification accuracy over all tissues with respect to ground truth, ii) the accuracy of the simulated electromagnetic wave propagation through the head, and iii) the accuracy of the image reconstruction of the hemorrhage. The segmentation accuracy was measured in terms of the degree of overlap (Dice score) with the ground truth. The electromagnetic simulation accuracy was measured in terms of signal deviation relative to the simulation based on the ground truth. Finally, the image reconstruction accuracy was measured in terms of the Dice score, relative error of dielectric properties, and visual comparison between the true and reconstructed intracerebral hemorrhage. The results show that accurate segmentation of tissues (Dice score = 0.97) from the MRI data can lead to accurate image reconstruction (relative error = 0.24) for the intracerebral hemorrhage in the subject’s brain. They also suggest that accurate automated segmentation can be used as a surrogate for manual segmentation and can facilitate the rapid diagnosis of intracerebral hemorrhage in stroke patients using a microwave imaging system.
Keywords:
Magnetic Resonance Imaging, Automatic Segmentation, Microwave, Dielectric Head Model, Intracerebral Hemorrhage Reconstruction

1. Introduction
Worldwide, about 15 million people have a stroke each year and a third of these die [1] . Stroke is a disturbance to the intracerebral blood flow caused by either a clot blocking an artery (ischemic stroke) or bleeding as the result of a burst blood vessel (hemorrhagic stroke) [2] . As a consequence, the oxygen and glucose levels are reduced which in turn rapidly kills the affected brain tissue; e.g. an estimated loss of almost 2 million neurons per minute takes places for a large-vessel ischemic stroke [3] . Ischemic stroke is the most common type of stroke accounting for about 80% of cases (the proportion varies between countries) [4] . For ischemic stroke patients, thrombolytic treatment that dissolves the clot is very effective if given shortly after onset [2] . Unfortunately, only 1% - 8% of ischemic stroke patients today receive thrombolytic treatment within the necessary time frame (4.5 h) because of delays in seeking medical attention and making a diagnosis [2] . Before thrombolytic treatment can be given, intracerebral hemorrhage has to be ruled out because treatment may aggravate the bleeding causing further brain damage and even death.
Computerized tomography (CT) is currently the gold standard for stroke diagnosis, with MRI often being used when CT does not give a definitive answer [5] . The main drawback of these imaging modalities is that they are bulky and not portable. Therefore, stroke patients have to be transported to the hospital to be diagnosed. Although purpose-built large ambulances with CT have been trialed with promising results [6] , this is an expensive solution and can only be considered in densely populated areas with well-developed traffic and telecommunications infrastructure. Ultrasound is a possible future portable alternative but at present is not effective for detecting intracerebral hemorrhage [7] . Thus there is a need for a fast, cost-effective and portable stroke detection imaging system that can be installed in ordinary ambulances.
To this end several microwave systems [8] -[11] have been proposed in recent years. The systems proposed in [8] - [10] were evaluated using numerical simulations whilst that proposed in [11] was used to obtain clinical measurements from stroke patients. Microwave imaging typically uses several microwave antennas positioned around the patient’s head and measures the amplitude and phase for the propagated signal between all possible pairs of antenna combinations. Typically either a single frequency is used or multiple frequencies in an interval around 1 GHz are used. The dielectric properties of the brain tissues govern the microwave scattering [12] . The high contrast between blood and brain tissue means that a bleeding can be readily detected. Microwave imaging is based on reconstructing dielectric images that can be related to physiological changes in the brain.
At present, full 3D reconstruction algorithms typically need several hours to reconstruct an image. This is not feasible in ambulances where rapid reconstructions are needed. In principle fast reconstructions can be obtained using a patient-specific dielectric model of the head obtained from an MRI scan obtained prior to the patient suffering a stroke. An image reconstruction can then be calculated as an update to the prior head model, wherein the measured signals from the patient’s head after they have suffered a stroke are compared with the corresponding simulations of the head model to localize changes in the tissue associated with the stroke.
The quality and fidelity of the dielectric head model necessarily impact on the accuracy of image reconstruction. The construction of a realistic dielectric head model involves, as a first step, the segmentation (delineation/ labeling) of the patient’s head tissues from MRI data obtained before the patient suffered a stroke. The segmentation is then used to construct a dielectric head model by assigning to each tissue type its corresponding dielectric property (conductivity/permittivity) value.
Accurate segmentation of brain tissues from MRI data is a challenging task because of several factors including: i) the complexity and variability of the underlying anatomy; ii) noise; iii) the bias field (an unwanted low- frequency signal caused by inhomogeneities in the magnetic fields of the MRI scanner); and iv) the low contrast between the skull, cerebrospinal fluid (CSF) and air in T1-weighted MRI data. Expert manual segmentation is today the gold standard but is time consuming, labor intensive and tedious.
In this paper we present a comparative study of several automated unsupervised (do not require training examples) segmentation algorithms for use in constructing a patient specific dielectric head model. The main purpose was to investigate how segmented head models could be used in a fast algorithm for reconstruction of hemorrhagic stroke. Another purpose was to investigate how the accuracy of the segmented model influenced the accuracy of the reconstructed image.
The algorithms were evaluated both directly in terms of segmentation accuracy and indirectly in terms of electromagnetic simulation accuracy and image reconstruction accuracy in a microwave tomography system for imaging intracerebral hemorrhage. The segmentation methods include the commonly used BET-FAST (brain extraction tool-FMRIB’s automated segmentation tool) [13] [14] method, and five variations on a hierarchical segmentation approach (HSA) wherein the MRI data are initially partitioned into brain and non-brain, followed by a second step where individual tissues are segmented using either Bayesian adaptive mean-shift (BAMS), adaptive mean-shift (AMS), k means, fuzzy c-means, or FAST. The study was performed using both synthetic MRI data and real data from four healthy subjects. In the case of the synthetic data, the labeled tissues used to generate the MRI data were used as “ground truth” segmentation. In the case of the real data, expert manual segmentation served as “ground truth”. The segmentations were done on the full 3D data. For computational reasons, the electromagnetic simulation and image reconstructions were done on a single 2D slice (located between the upper head and the eyes) from the 3D segmented MRI data.
The simulations were performed using a model of a microwave imaging system [15] , consisting of 20 antennas, placed equidistantly around a 2D dielectric head model (constructed from the 2D segmented slice). Scattering measurements (amplitude and phase of the simulated scattering signals) were obtained from a dielectric head model constructed from each segmentation method independently, and for a dielectric head model constructed from the ground truth with a simulated intracerebral hemorrhage. The scattering measurements were used in the image reconstruction algorithm.
2. Materials and Methods
2.1. Proposed Stroke Diagnosis Procedure
This section describes the proposed stroke diagnosis procedure using the microwave imaging system (see Figure 1). It is assumed that brain MRI data exists for the patient prior to the onset of stroke; e.g. this might be routinely done for high-risk patients. The MRI data (Figure 1, top-left) is segmented (see Section 2.3) to construct a patient specific dielectric head model (Figure 1, bottom-left). This model is then used to simulate the scattering parameters (transmission and reflection coefficients) expected from the actual microwave measuring system (see Section 2.4).
The stroke patient is measured with the microwave imaging system (Figure 1, top-right). The simulated and measured scattering parameters are then compared (Figure 1, center-right) in the image reconstruction algorithm (see Section 2.4.1), wherein the reconstruction is generated based on the difference between these parameters. This reconstruction (Figure 1, bottom-right) shows the change in dielectric properties of the brain tissues associated with bleeding. Both the measurement and reconstruction are designed to be performed in an ambulance equipped with a microwave based stroke diagnostic system. Note that for the purposes of this study, we mimic this scenario by using simulations of an intracerebral bleeding in place of real measurements.
2.2. MRI Data
Synthetic multi-modal MRI data (i.e. data acquired using different MRI techniques) together with real multi- modal MRI data from four different healthy subjects were used in this study. These data are hereinafter referred to as the synthetic dataset, real dataset 1, real dataset 2, real dataset 3, and real dataset 4 respectively.
The synthetic dataset comprises T1-weighted, T2-weighted, and proton density-weighted (PD) MRI data, for 5% noise level and 20% bias field level, obtained from the BrainWeb simulated brain database [16] .
Figure 1. Schematic of the proposed stroke diagnosis procedure using the microwave imaging system.
Real dataset 1 comprises T1- and T2-weighted images of the head of a healthy subject. The data were acquired on a 3T (Tesla) Philips Achieva scanner.
Real datasets 2 - 4 comprise T1-, T2- and PD-weighted scans of the heads of three healthy subjects respectively from the IXI dataset (IXI040_Guys_0724, IXI121_Guys_0772, and IXI144_Guys_0788) [17] . The data were acquired using a Philips Medical Systems Intera 1.5T scanner. Details of the scan parameters and image and voxel dimensions for each MRI dataset are shown in Appendix (Table A1).
2.3. Segmentation Methods
2.3.1. Manual Segmentation (Ground Truth)
In the case of the synthetic MRI dataset the nine tissue classes―white matter, gray matter, CSF, skull, skin, fat, muscle, connective, and glial matter―defined in the BrainWeb simulated brain database [16] were taken to be ground truth segmentation. This was reduced to seven classes by merging tissues with similar dielectric properties. In particular the connective and fat tissue classes were merged, and the glial matter and gray matter classes were merged.
For the real MRI datasets manual segmentations, by an experienced radio-oncologist, into five tissue classes- white matter, gray matter, CSF, skull and skin―were taken as the ground truth. The decision to use five tissue classes represented a trade-off between the time needed to manually segment the images and the number of tissue classes. The manual segmentation took about 170 hours for 200 slices.
2.3.2. Automated Segmentation Methods
Six automatic (unsupervised) segmentation methods were compared in this study for use in constructing a dielectric head model of a subject, from MRI data acquired before the stroke event. Five are variations on a hierarchical segmentation approach and the last is a combination of the BET and FAST tools from the FMRIB Software Library (FSL).
1) Hierarchical Segmentation Approach
A schematic of the hierarchical segmentation approach (HSA) is presented in Figure 2. It takes as input multi-modal spatially-aligned MRI data of a subject (Figure 2, top-left). This data can be modeled as a single spatial head volume (V) with vector-valued voxels. In the first level of the HSA (marked level 1 in Figure 2), the head volume (V) is segmented into brain and non-brain tissue. For this the T1-weighted data is used to obtain both a brain mask and a whole head mask (Figure 2, top-center). The BET (Brain Extraction Tool) [13] is used to obtain the former and a simple whole head segmentation (WHSA) algorithm [18] [19] is used to obtain the latter. The set difference (“/”) between these two masks then yields a mask of the non-brain head tissue (Figure 2, top- right). These masks effectively partition the head volume (V) into two disjoint sub-volumes: brain tissue (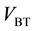 ) and non-brain tissue (
) and non-brain tissue (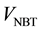 ) (Figure 2, center). In the second level of the HSA (marked level 2 in Figure 2), the brain and non-brain tissue are segmented into their individual tissue classes. This is achieved by applying a multi-tissue segmentation algorithm (MTSA) independently to the brain and non-brain tissue. The brain tissue is segmented into white matter, gray matter, and CSF tissue (Figure 2, bottom image on left side) while the non- brain tissue is segmented into skin, skull, fat, and muscle tissue or skin, skull tissue (Figure 2, bottom image on right side).
) (Figure 2, center). In the second level of the HSA (marked level 2 in Figure 2), the brain and non-brain tissue are segmented into their individual tissue classes. This is achieved by applying a multi-tissue segmentation algorithm (MTSA) independently to the brain and non-brain tissue. The brain tissue is segmented into white matter, gray matter, and CSF tissue (Figure 2, bottom image on left side) while the non- brain tissue is segmented into skin, skull, fat, and muscle tissue or skin, skull tissue (Figure 2, bottom image on right side).
2) Multi-Tissue Segmentation Algorithms (MTSAs)
We used five different unsupervised multi-tissue segmentation algorithms (MTSAs): adaptive mean shift (AMS) [20] , Bayesian adaptive mean shift (BAMS) [21] , FMRIB’s automated segmentation tool (FAST) [14] , k means [22] , and fuzzy c-means [23] , independently in the hierarchical segmentation approach. AMS and BAMS (see [20] and [21] respectively for more detail) are variations on the mean shift algorithm. Mean shift [24] [25] is based on an adaptive gradient ascent approach to estimate the local maxima or modes of the probability density function underlying the feature space (features could be colors, texture, etc.). Ultimately each feature point is associated with a mode (local maxima of the probability density function corresponding to a dense region in the feature space) thereby defining clusters (groups of features points). The advantages of mean shift are that it doesn’t require any initialization for clustering and performs well in the presence of noise and the bias field.
Figure 2. Schematic of the hierarchical segmentation approach (HSA) for automated whole head segmentation. In the figure, MTSA stands for multi- tissue segmentation algorithm.
The FAST tool [14] is commonly used for brain tissue segmentation. The underlying method is based on the hidden Markov random field model and associated Expectation-Maximization (HMRF-EM) algorithm [14] .
The methods k means [22] and fuzzy c-means [23] are based on the distance function to partition the input data into clusters. These methods are frequently used in the pattern recognition field.
Hereinafter, the five variations of the HSA are denoted HSA-AMS, HSA-BAMS, HSA-FAST, HSA-k means and, HSA-fuzzy c-means.
3) FMRIB Software Library (FSL) tools: BET-FAST
The combination of the BET [13] and FAST [14] tools (BET-FAST) in FSL is commonly used for the creation of patient-specific dielectric models [26] - [28] . The BET-FAST segmentation approach takes the same inputs as the hierarchical segmentation approach (HSA), shown in Figure 2. The BET tool is applied to extract the brain- tissue mask, skull-tissue mask, and skin-tissue mask from the T1-weighted images (similar to HSA level 1 in Figure 2). The FAST tool is then used to segment the brain-only image (masked version of the T1-weighted images) into three tissue classes: white matter, gray matter, and CSF (similar to HSA level 2 in Figure 2).
2.3.3. Optimal Parameters Settings for the MTSAs and BET-FAST tool
In this study, the AMS and BAMS MTSAs were applied with the same parameter settings as described in [20] and [21] respectively. The k means, fuzzy c-means, and FAST MTSAs were initialized using the prior knowledge of tissue intensity ordering in the T1-weighted MRI scan. For example, in the T1-weighted MRI scan the highest intensity value was used to initialize the cluster for WM tissue, the lowest intensity value was used to initialize the cluster for CSF tissue, and the mean of the highest and lowest intensity values was used to initialize the cluster for GM tissue. Moreover, for the fuzzy c-means, the parameter m (controls the fuzziness of the resulting clusters) was set to 1.5 and for FAST, the parameter H (MRF beta) was set to 0.1. For the brain, skull and skin mask extraction, the BET tool was applied using the threshold parameter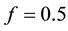 .
.
2.3.4. Computational Complexity and Time
Since the mean shift is an iterative intensive algorithm, the methods: HSA-AMS and HSA-BAMS have high computational complexity compared to the other competing methods. The approximate execution time for each method for the segmentation of the synthetic multi-modal 181 × 217 × 181 MRI head volume on a single desktop PC running 64-bit Ubuntu 10.04 with 8 GiB ram memory and Intel Core i7 CPU 870 at 2.93 GHz is presented in Table A2 (see Appendix).
2.4. Procedure for Evaluating the Intracerebral Hemorrhage Reconstruction
In this study, the MRI datasets were independently segmented in 3D using each segmentation method. A single 2D axial slice (slice between the upper head and the eyes) from each segmented dataset was then independently used to construct a 2D dielectric head model (representing the situation where there is no hemorrhage) by assigning to each tissue class conductivity/permittivity properties obtained from [12] . A dielectric head model was similarly constructed from the ground truth segmentation for each MRI data set and manually modified to yield dielectric head models corresponding to intracerebral hemorrhages of different sizes and locations. Eight different hemorrhages were simulated by superimposing two different sized disks in four different positions (see Figure 3). The radii for the large and small intracerebral hemorrhages were 20 mm and 10 mm, respectively.
Each 2D dielectric head model without intracerebral hemorrhage was used in an electromagnetic simula- tion to compute scattering parameters (see Figure 1, bottom-left side). In principle these parameters can be
Figure 3. Dielectric head model with a large simulated intracerebral hemorrhage (a)-(d) and with a small simulated intracerebral hemorrhage (e)-(h) for four different positions.
pre-computed to facilitate real time reconstruction. Scattering parameters were similarly obtained for each dielectric head model corresponding to a hemorrhage (in the real world these would be obtained from the stroke patient using a helmet fitted with a microwave antenna array).
The antenna configuration for the microwave imaging system, used in this study for electromagnetic simulations, is shown in Figure 4. It comprises 20 antennas placed equidistantly around the dielectric head model. The antennas are used both as transmitters and receivers. For each tomographic measurement, one antenna at the time is used as transmitter while the others are used as receivers to measure the scattered signals. This process is continued until all antennas have been used as transmitters and all possible antenna combinations have been measured.
The electromagnetic simulations were performed with a 2D finite-difference time domain (FDTD) method [29] [30] . The FDTD method was used to simulate the electric fields inside the head as well as to simulate the scattering parameters from the head. The electric fields and scattering parameters for each pair comprising a head model without hemorrhage and the corresponding head model with a simulated hemorrhage were then used as inputs to the image reconstruction algorithm in order to reconstruct the intracerebral hemorrhage (see Figure 1, bottom-right).
Reconstruction Algorithm
The algorithm takes as input the simulated and measured signals, obtained from the head model without and the corresponding head model with a simulated hemorrhage respectively, and processes this data in order to determine the change in dielectric properties of the brain tissues associated with the intracerebral hemorrhage. In this study, we used a fast reconstruction algorithm, which is based on a Born approximation [31] , wherein it is assumed that the scattering electric field due to the small perturbation (intracerebral hemorrhage) is negligible compared to the incident electric field.
Based on the Born approximation, the difference between the scattering parameters (obtained from the FDTD simulation as described above) simulated using the dielectric head model without hemorrhage and the scattering parameters simulated using the dielectric head model with hemorrhage (in practice, measured) [32] [33] is given by
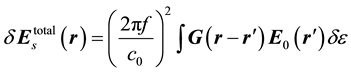 (1)
(1)
where  represents the change in dielectric properties of the brain tissues due to the intracerebral hemorrhage,
represents the change in dielectric properties of the brain tissues due to the intracerebral hemorrhage,  is the incident electric field in the image domain and G is the Green’s function that represents the electric field resulting from the pixels acting as single point scatterers in the image domain [34] . Both
is the incident electric field in the image domain and G is the Green’s function that represents the electric field resulting from the pixels acting as single point scatterers in the image domain [34] . Both 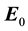 and G are obtained using the dielectric head model without hemorrhage. For simplicity, Equation 1 can be expressed as
and G are obtained using the dielectric head model without hemorrhage. For simplicity, Equation 1 can be expressed as
Figure 4. Configuration of the microwave imaging system for electromagnetic simulations (the figure shows an example 2D dielectric head model with simulated intracerebral hemorrhage).
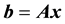 (2)
(2)
where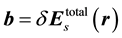 ,
, 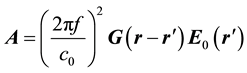 , and
, and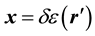 .
.
In order to construct the intracerebral hemorrhage, we solved Equation (2) for . Equation (2) represents a system of ill-posed linear equations. To make it well posed, a Tikhonov regularization was used [35] . Equation (2) then becomes
. Equation (2) represents a system of ill-posed linear equations. To make it well posed, a Tikhonov regularization was used [35] . Equation (2) then becomes

where 

The solution of Equation (3) is then

where 

2.5. Quantitative Evaluation
The quantitative evaluation of each segmentation method for each data was done in terms of segmentation accuracy over all tissues, electromagnetic simulation accuracy, and reconstruction image accuracy for intracerebral hemorrhage.
Segmentation accuracy for each tissue type, each method and each dataset was evaluated in terms of the Dice score [36] . The Dice score measures the degree of overlap between the ground truth and the segmentation. The Dice score for the ith tissue is given by

where 



where N is the total number of tissues.
The electromagnetic simulation accuracy was evaluated in terms of signal deviation (F). The signal deviation was defined as the sum of squared difference between the scattering signals obtained from the dielectric models with and without intracerebral hemorrhage, respectively. It is given by

where N is the total number of antennas, n is the total number of signal samples and 

The reconstruction image accuracy for both conductivity and permittivity of intracerebral hemorrhage for each dataset and each segmentation method was measured with respect to the ground truth in terms of relative error.
The relative error (RE) for the permittivity [29] is given by

where S is the region of intracerebral hemorrhage being reconstructed, εGT represents the permittivity profile of the intracerebral hemorrhage reconstruction using the ground truth, and εASM is the permittivity profile of the intracerebral hemorrhage reconstruction using the automatic segmentation method. The relative error for the conductivity is similarly defined.
We also measured the accuracy of image reconstruction in terms of the Dice score by computing the degree of overlap between the actual (simulated bleeding in the ground truth brain) and reconstructed intracerebral hemorrhage.
Finally the mean signal deviation, mean relative error and mean Dice score were computed over the four different positions for both large and small intracerebral hemorrhages (see Figure 3).
Optimal Parameters Settings for the Reconstruction Algorithm
The ground truth from the synthetic data was used to empirically determine the optimal settings for the 



We have previously shown that different sized objects are best reconstructed at different frequencies [30] . Therefore we have used different frequencies when reconstructing the large and the small intracerebral hemorrhage respectively. For the large hemorrhages f = 350 MHz was used. For the small hemorrhages separate reconstructions were made at f = 350, 550, 750 and 850 MHz. Averaging all the individual reconstructed dielectric images then generated the final reconstruction. The optimal value of α was empirically found to be α = 0.05.
3. Results and Discussion
3.1. Qualitative Evaluation for Segmentation Accuracy
Figure 5 shows a segmentation example for each method including ground truth for a single axial slice from the synthetic dataset for 5% noise level with 20% bias field level. It can be seen that the methods HSA-BAMS and HSA-AMS yield fewer segmentation errors (every misclassified pixel is shown with a black dot) than the other methods. This suggests that HSA-BAMS and HSA-AMS are more robust to the noise and bias field. The methods: HSA-k means, HSA-fuzzy c-means, HSA-FAST and BET-FAST have a higher tendency to misclassify the gray matter as CSF. Moreover, HSA-k means and HSA-fuzzy c-means have a higher tendency to misclassify the fat as muscle tissue and the muscle as skin tissue while HSA-FAST has a higher tendency to misclassify the fat as muscle and skin tissue. However, in the BET-FAST method, BET is limited to segmenting the non-brain into two tissue types: skin and skull, wherein the fat and muscle are included in the “skin” class. Therefore,


Figure 5. Segmentation results for the synthetic dataset for each segmentation method. (a) Ground truth; (b) HSA-BAMS; (c) HSA-AMS; (d) HSA-k means; (e) HSA-fuzzy c-means; (f) HSA-FAST; (g) BET-FAST.
BET-FAST has higher misclassification of skin tissue. Moreover, the BET-FAST method over segments the skull tissue.
Figure 6 shows a segmentation example for each method for a single axial slice from real dataset 2. It can be seen that the method HSA-BAMS yields fewer segmentation errors (shown in black dot) than the other methods. The methods: HSA-k means, HSA-fuzzy c-means, HSA-FAST and BET-FAST have a higher tendency to misclassify the white matter as gray matter and the gray matter as CSF. Moreover HSA-AMS as well as other competing methods (HSA-k means, HSA-fuzzy c-means. HSA-FAST and BET-FAST) over segment the skull tissue. For the other real datasets (dataset 1, dataset 3 and dataset 4), similar axial slices were selected for the evaluation and the qualitative results for these datasets were similar to those of dataset 2 (shown in Figure 6).
3.2. Segmentation Accuracy versus Electromagnetic Microwave Simulation Accuracy
The accuracy of image reconstruction depends on the accuracy of electromagnetic microwave simulation. In turn the accuracy of the electromagnetic simulation depends on the segmentation accuracy. Combined plots of segmentation accuracy (Dice score over all tissues) and electromagnetic simulation accuracy (signal deviation over all simulated hemorrhages) are shown in Figure 7 for each segmentation method and each dataset.
The results in Figure 7 show that the method HSA-BAMS has the lowest mean signal deviation and highest mean Dice score compared to the other methods. The results also show that the mean signal deviation measured using only the ground truth is very small compared to that measured using the automated segmentation methods. This suggests that tissue segmentation accuracy affects the simulated scattering signals more than the intracerebral hemorrhage.
3.3. Qualitative Evaluation for Intracerebral Hemorrhage Reconstruction
Example reconstructions of the permittivity and conductivity profiles for a large intracerebral hemorrhage with radius size of 20 mm from the synthetic dataset are shown in Figure 8 and Figure 9 respectively.
Example reconstructions of the permittivity and conductivity profiles for a small intracerebral hemorrhage with radius size of 10 mm from the real dataset 2 are shown in Figure 10 and Figure 11 respectively.
In each figure, the contours in the reconstructed images show the structures of the head tissues, wherein the actual intracerebral hemorrhage is shown as a circular contour. The color bar represents the range of intensity for the reconstructed images. In the reconstructed images, the white pixels correspond to background and the yellow and red pixels correspond to the intracerebral hemorrhage.
Figure 8 and Figure 9 show that the reconstructions of the large intracerebral hemorrhage obtained using the
Figure 6. Segmentation results for the real dataset 2 for each segmentation method. (a) Ground truth; (b) HSA-BAMS; (c) HSA-AMS; (d) HSA-k means; (e) HSA-fuzzy c-means; (f) HSA-FAST; (g) BET-FAST.


Figure 7. Mean Dice score and mean signal deviation (in logarithmic scale) for (a) large (b) small intracerebral hemorrhage. Results for different segmentation methods are shown along the x-axis, for every method a group of data is shown with different colors, corresponding to different datasets. Along the positive y-axis the histograms show the mean Dice score and on the negative y-axis the mean signal deviation in logarithmic scale.

Figure 8. Reconstruction of the permittivity profile for a large intracerebral hemorrhage with radius size of 20 mm from the synthetic dataset obtained using the different segmentation methods. (a) Ground truth; (b) HSA-BAMS; (c) HSA-AMS; (d) HSA-k means; (e) HSA-fuzzy c-means; (f) HSA-FAST; (g) BET-FAST.

Figure 9. Reconstruction of the conductivity profile for a large intracerebral hemorrhage with radius size of 20 mm from the synthetic dataset obtained using the different segmentation methods. (a) Ground truth; (b) HSA-BAMS; (c) HSA-AMS; (d) HSA-k means; (e) HSA-fuzzy c-means; (f) HSA-FAST; (g) BET-FAST.
segmentation methods HSA-BAMS and HSA-AMS are better than that obtained using the HSA-k means, HSA- fuzzy c-means, HSA-FAST and the BET-FAST method, and are similar to that obtained using the ground truth.
Figure 10 and Figure 11 show that the reconstructions of the small intracerebral hemorrhage obtained using the segmentation method HSA-BAMS are better than that obtained using the competing methods and are similar to that obtained using the ground truth. Figure 10 also shows that there is no overlapping between the location of the actual intracerebral hemorrhage and the location of the reconstructed intracerebral hemorrhage obtained using HSA-k means, HSA-fuzzy c-means, HSA-FAST and the BET-FAST method.
3.4. Quantitative Evaluation for Intracerebral Hemorrhage Reconstruction
The reconstructed images for both normalized permittivity and conductivity are shown in Figures 8-11. In order to quantitatively assess the accuracy of the reconstructions, the first problem is to define what part of the reconstructed object defines the boundaries of the bleeding. It is not immediately straightforward how to do this and quantifying the levels of artifacts and reconstructions would require further investigations. We have therefore taken the following approach in the analysis, a threshold level “T” for both normalized permittivity and conductivity was defined. It was used to discriminate the intracerebral hemorrhage from the background. To exemplify, three different values T = 0.6, 0.7, 0.8 were selected and used for further analysis. The images were normalized to have maximum value 1, the permittivity/conductivity values below the threshold level, T were treated as background while the permittivity/conductivity values above T were considered to represent the intracerebral hemorrhage. The Dice score was then measured between the actual and reconstructed intracerebral hemorrhage for both permittivity and conductivity.
Figure 12 shows the combined plots for the mean Dice score for the threshold level 0.7 and the mean relative error for permittivity for both large and small intracerebral hemorrhages.
The plots for the synthetic data for both large and small intracerebral hemorrhages show that the methods HSA-BAMS and HSA-AMS have better reconstruction accuracy (higher mean Dice score and lower relative error) than HSA-k means, HSA-fuzzy c-means, HSA-FAST and BET-FAST. The plots for each real dataset show that, relative to the ground truth, HSA-BAMS has better reconstruction accuracy (higher mean Dice score and lower relative error) than all other segmentation methods. For the small intracerebral hemorrhage, it can also be observed that HSA-k means, HSA-fuzzy c-means, HSA-FAST and BET-FAST have zero mean Dice score; this indicates that there is no overlap between the reconstructed intracerebral hemorrhage and the actual intracerebral hemorrhage. This can be clearly seen in Figures 10(d)-(g).
In the case of conductivity, similar results were obtained for the mean Dice score and mean relative error for both large and small intracerebral hemorrhages.

Figure 10. Reconstruction of the permittivity profile for a small intracerebral hemorrhage with radius size of 10 mm from the real dataset 2 obtained using the different segmentation methods. (a) Ground truth; (b) HSA-BAMS; (c) HSA-AMS; (d) HSA-k means; (e) HSA-fuzzy c-means; (f) HSA-FAST; (g) BET-FAST.

Figure 11. Reconstruction of the conductivity profile for a small intracerebral hemorrhage with radius size of 10 mm from the real dataset 2 obtained using the different segmentation methods. (a) Ground truth; (b) HSA-BAMS; (c) HSA-AMS; (d) HSA-k means; (e) HSA-fuzzy c-means; (f) HSA-FAST; (g) BET-FAST.


Figure 12. Mean Dice score and mean relative error for permittivity for (a) large and (b) small intracerebral hemorrhage for each method and each data.
The mean Dice score was also computed for the other threshold levels 0.6 and 0.8 respectively. For each method, similar trends were found in the results of permittivity and conductivity for both large and small intracere bral hemorrhages.
4. Conclusions
In this study, we presented an evaluation of six automatic segmentation methods for use in the construction of a dielectric head model for imaging an intracerebral hemorrhage in MR brain images using a simulated microwave imaging system. The evaluation was performed using synthetic MRI data as well as real MRI data from four different healthy subjects. The evaluation was done in terms of segmentation accuracy over all tissues, electromagnetic simulation accuracy, and image reconstruction accuracy for the intracerebral hemorrhage in the subject’s brain for four different positions with two different sizes.
The results show that the automatic segmentation method HSA-BAMS has better performance (higher segmentation accuracy, lower signal deviation and lower relative error) compared with the other methods. The results also indicate that accurate segmentation of tissues leads to accurate reconstruction of intracerebral hemorrhage in the subject’s brain. Overall the results suggest that automated segmentation (in particular HSA-BAMS) can be used as a surrogate for manual segmentation of the head tissues required for the image reconstruction algorithm.
This study represents a first step towards the visualization of intracerebral hemorrhage for stroke patients in near real-time using contemporary computing power in mobile devices suitable for field use. In the future, we aim to investigate the performance of the automatic segmentation method HSA-BAMS for the reconstruction of intracerebral hemorrhage in MR images using microwave-based measurements from stroke patients.
Acknowledgements
The author Mr. Mahmood acknowledges scholarship funding from the Higher Education Commission of Pakistan (HEC) and Chalmers University of Technology in support of this work.
References
- The Atlas of Heart Disease and Stroke. http://www.who.int/cardiovascular_diseases/resources/atlas/en/
- Fassbender, K., Balucani, C., Walter, S., Levine, S.R., Haass, A. and Grotta, J. (2013) Streamlining of Prehospital Stroke Management: The Golden Hour. Lancet Neurology, 12, 585-596. http://dx.doi.org/10.1016/S1474-4422(13)70100-5
- Burns, J.D., Green, D.M., Metivier, K. and DeFusco, C. (2012) Intensive Care Management of Acute Ischemic Stroke. Emergency Medicine Clinics of North America, 30, 713-744. http://dx.doi.org/10.1016/j.emc.2012.05.002
- Feigin, V.L., Lawes, C.M., Bennett, D.A., Barker-Collo, S.L. and Parag, V. (2009) Worldwide Stroke Incidence and Early Case Fatality Reported in 56 Population-Based Studies: A Systematic Review. Lancet Neurology, 8, 355-369. http://dx.doi.org/10.1016/S1474-4422(09)70025-0
- Chalela, J., Kidwell, C.S., Nentwich, L.M., Luby, M., Butman, J.A., et al. (2007) Magnetic Resonance Imaging and Computer Tomography in Emergency Assessment of Patients with Suspected Acute Stroke: A Prospective Comparison. Lancet, 369, 293-298. http://dx.doi.org/10.1016/S0140-6736(07)60151-2
- Walter, S., Kostopoulos, P., Haass, A., Keller, I., Lesmeister, M., et al. (2012) Diagnosis and Treatment of Patients with Stroke in a Mobile Stroke Unit versus in Hospital: A Randomised Controlled Trial. Lancet Neurology, 11, 397- 404. http://dx.doi.org/10.1016/S1474-4422(12)70057-1
- Hölscher, T., Dunford, J.V., Schlachetzki, F., Boy, S., Hemmen, T., et al. (2013) Prehospital Stroke Diagnosis and Treatment in Ambulances and Helicopters―A Concept Paper. American Journal of Emergency Medicine, 31, 743-747. http://dx.doi.org/10.1016/j.ajem.2012.12.030
- Ireland, D. and Bialkowski, M. (2011) Microwave Head Imaging for Stroke Detection. Progress in Electromagnetics Research M, 21, 163-175. http://dx.doi.org/10.2528/PIERM11082907
- Scapaticci, R., Di Donato, L., Catapano, I. and Crocco, L. (2012) A Feasibility Study on Microwave Imaging for Brain Stroke Monitoring. Progress in Electromagnetics Research B, 40, 305-324. http://dx.doi.org/10.2528/PIERB12022006
- Irishina, N. and Torrente, A. (2013) Brain Stroke Detection by Microwaves Using Prior Information from Clinical Databases. Abstract and Applied Analysis, 2013, Article ID: 412638. http://dx.doi.org/10.1155/2013/412638
- Persson, M., Fhager, A., Trefna, H.D., Yu, Y.N., McKelvey, T., Pegenius, G., et al. (2014) Microwave-Based Stroke Diagnosis Making Global Prehospital Thrombolytic Treatment Possible. IEEE Transactions on Biomedical Engineering, 61, 2806-2817. http://dx.doi.org/10.1109/TBME.2014.2330554
- Dielectric Properties of Body Tissues. http://niremf.ifac.cnr.it/tissprop/
- Smith, S.M. (2002) Fast Robust Automated Brain Extraction. Human Brain Mapping, 17, 143-155. http://dx.doi.org/10.1002/hbm.10062
- Zhang, Y., Brady, M. and Smith, S. (2001) Segmentation of Brain MR Images through a Hidden Markov Random Field Model and the Expectation Maximization Algorithm. IEEE Transactions on Medical Imaging, 20, 45-57. http://dx.doi.org/10.1109/42.906424
- Fhager, A. and Persson, M. (2007) Using a Priori Data to Improve the Reconstruction of Small Objects in Microwave Tomography. IEEE Transactions on Microwave Theory and Techniques, 55, 2454-2462. http://dx.doi.org/10.1109/TMTT.2007.908670
- Cocosco, C.A., Kollokian, V., Kwan, R.K.S. and Evans, A.C. (1997) BrainWeb: Online Interface to 3-D MRI Simulated Brain Database. NeuroImage, 5, S425.
- IXI Dataset. http://www.brain-development.org/
- Otsu, N. (1979) A Threshold Selection Method from Gray-Level Histrogram. IEEE Transactions on Systems, Man, and Cybernetics, 9, 62-66. http://dx.doi.org/10.1109/TSMC.1979.4310076
- Soille, P. (1999) Morphological Image Analysis: Principles and Applications. Springer-Verlag, Berlin, 173-174. http://dx.doi.org/10.1007/978-3-662-03939-7
- Mayer, A. and Greenspan, H. (2009) An Adaptive Mean-Shift Framework for MRI Brain Segmentation. IEEE Transactions on Medical Imaging, 28, 1238-1249. http://dx.doi.org/10.1109/TMI.2009.2013850
- Mahmood, Q., Chodorowski, A., Mehnert, A. and Persson, M. (2012) A Novel Bayesian Approach to Adaptive Mean Shift Segmentation of Brain Images. Proceedings of the IEEE International Symposium on Computer-Based Medical Systems (CBMS), Rome, 20-22 June 2012, 1-6. http://dx.doi.org/10.1109/CBMS.2012.6266304
- Seber, G.A.F. (1984) Multivariate Observations. John Wiley & Sons, Inc., Hoboken. http://dx.doi.org/10.1002/9780470316641
- Bezdek, J.C. (1981) Pattern Recognition with Fuzzy Objective Function Algorithms. Kluwer Academic Publishers, Norwell. http://dx.doi.org/10.1007/978-1-4757-0450-1
- Comaniciu, D., Ramesh, V. and Meer, P. (2001) The Variable Bandwidth Mean-Shift and Data-Driven Scale Selection. Proceedings of the IEEE International Conference on Computer Vision (ICCV), Vancouver, 7-14 July 2001, 438-445. http://dx.doi.org/10.1109/ICCV.2001.937550
- Comaniciu, D. and Meer, P. (2002) Mean Shift: A Robust Approach toward Feature Space Analysis. IEEE Transactions on Pattern Analysis and Machine Intelligence, 24, 603-619. http://dx.doi.org/10.1109/34.1000236
- Shirvany, Y., Edelvik, F., Jakobsson, S., Hedström, A., Mahmood, Q., Chodorowski, A. and Persson, M. (2012) Non- Invasive EEG Source Localization Using Particle Swarm Optimization: A Clinical Experiment. Proceedings of the 34th Annual International Conference of the IEEE EMBS, San Diego, 28 August-1 September 2012, 6232-6235.
- Rullmann, M., Anwander, A., Dannhauer, M., Warfield, S.K., Duffy, F. and Wolters, C. (2009) EEG Source Analysis of Epileptiform Activity Using a 1 mm Anisotropic Hexahedra Finite Element Head Model. NeuroImage, 44, 399-410. http://dx.doi.org/10.1016/j.neuroimage.2008.09.009
- Lanfer, B., Scherg, M., Dannhauer, M., Knösche, T.R., Burger, M., Wolters, C.H. (2012) Influences of Skull Segmentation Inaccuracies on EEG Source Analysis. NeuroImage, 62, 418-431. http://dx.doi.org/10.1016/j.neuroimage.2012.05.006
- Fhager, A. (2006) A Microwave Tomography. Doktorsavhandlingar vid Chalmers tekniska högskola, Gothenburg.
- Fhager, A., Hashemzadeh, P. and Persson, M. (2006) Reconstruction Quality and Spectral Content of an Electromagnetic Time-Domain Inversion Algorithm. IEEE Transactions on Biomedical Engineering, 53, 1594-1604. http://dx.doi.org/10.1109/TBME.2006.878079
- Semenov, S.Y., Bulyshev, A.E., Abubakar, A., Posukh, V.G., Sizov, Y.E., Souvorov, A.E., et al. (2005) Microwave- Tomographic Imaging of the High Dielectric-Contrast Objects Using Different Image-Reconstruction Approaches. IEEE Transactions on Microwave Theory and Techniques, 53, 2284-2294. http://dx.doi.org/10.1109/TMTT.2005.850459
- Slaney, M., Kak, A.C. and Larsen, L.E. (1984) Limitations of Imaging with First-Order Diffraction Tomography. IEEE Transactions on Microwave Theory and Techniques, 32, 860-874. http://dx.doi.org/10.1109/TMTT.1984.1132783
- Pahomov, V., Semenchik, V. and Kurilo, S. (2005) Reconstructing Reflecting Object Images Using Born Approximation. Proceedings of the IEEE 35th European Microwave Conference, Paris, 4-6 October 2005.
- Larsen, L.E. and Jacobi, J.H. (1986) Medical Applications of Microwave Imaging. IEEE Press, Piscataway.
- Tikhonov, A.N. and Arsenin, V.Y. (1977) Solutions of Ill-Posed Problems. Winston and Sons, Washington DC.
- Dice, L.R. (1945) Measures of the Amount of Ecologic Association between Species. Ecology, 26, 297-302. http://dx.doi.org/10.2307/1932409
Appendix
Table A1. Scan parameters and image and voxel dimensions for each MRI dataset.
Table A2. Approximate execution time for each segmentation method.







