American Journal of Molecular Biology
Vol.07 No.03(2017), Article ID:77807,15 pages
10.4236/ajmb.2017.73011
Identification and Characterization of Reverse Transcriptase Fragments of Long Interspersed Nuclear Elements (LINEs) in the Morus notabilis Genome
Bi Ma*, Youchao Xin*, Lulu Kuang, Fei Hou, Ningjia He#
State Key Laboratory of Silkworm Genome Biology, Southwest University, Chongqing, China

Copyright © 2017 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: May 1, 2017; Accepted: July 17, 2017; Published: July 21, 2017
ABSTRACT
Reverse transcriptase (rt) fragments from LINE retrotransposons in the mulberry genome were analyzed in terms of heterogeneity, phylogeny, and chromosomal distribution. We amplified and characterized conserved domains of the rt using degenerate primer pairs. Sequence analyses indicated that the rt fragments were highly heterogeneous and rich in A/T bases. The sequence identity ranged from 31.8% to 99.4%. Based on sequence similarities, the rt fragments were categorized into eight groups. Furthermore, similar stop codon distribution patterns among a series of clones in the same group indicated that they underwent a similar evolutionary process. Interestingly, phylogenetic analyses of the rt fragments isolated from mulberry and 13 other plant species revealed that two distantly related taxa (mulberry and Paeonia suffruticosa) grouped together. It does not appear that this phenomenon resulted from horizontal transposable element transfer. Fluorescence in situ hybridization analysis revealed that most of the rt fragments were concentrated in the subtelomeric and pericentromeric regions of the mulberry chromosomes, but that these elements were not abundant in the mulberry genome. Future studies will focus on the potential roles of these elements in the subtelomeric and pericentromeric regions of the mulberry genome.
Keywords:
Morus notabilis, LINE Retrotransposons, Reverse Transcriptase, Characterization, Fluorescence in site Hybridization

1. Introduction
Transposable elements (TEs), which were first discovered in maize by Barbara McClintock, are also known as “jumping genes” because of their ability to replicate and move to new genomic locations [1] . They are ubiquitous and abundant components of all eukaryotic genomes, and play important roles in the structural organization and evolution of genes and genomes [2] - [7] . Based on their mechanism of transposition, TEs are classified as retrotransposons (Class I) or DNA transposons (Class II) [8] [9] [10] . Class I retrotransposons move to new chromosomal locations via an RNA intermediate (i.e., “copy and paste” mechanism). In contrast, Class II DNA transposons move via a DNA intermediate (i.e., “cut and paste” mechanism) [6] [8] . Depending on whether or not they are flanked by long terminal repeats (LTRs), retrotransposons can be further classified as LTR or non-LTR retrotransposons [10] . Non-LTR retrotransposons are usually further divided into long or short interspersed nuclear elements (LINEs and SINEs, respectively) [10] .
The LINE retrotransposons are ubiquitous, showing a great variation in structure and size [11] [12] [13] . A large body of knowledge on mammalian LINEs has been accumulated [14] [15] . In contrast, LINEs in plants have been poorly investigated. The first identified plant LINE retrotransposon was Cin4 in Zea mays, which inactivates the A1 gene following its insertion into the A1 3'-untranslated region [16] . Since that pioneer study, numerous other LINEs have been identified in taxa such as Lilium speciosum (del2) [17] , Arabidopsis thaliana (Tal 1-1) [18] , Chlorella vulgaris (Zepp) [19] , Hordeum vulgare (BLIN) [20] , Oryza sativa (Karma) [21] , Ipomoea batatas (LIb) [22] , Beta vulgaris (BNR) [13] , and so on. Full-length LINEs have one or two open reading frames (ORFs) encoding proteins required for reverse transcription. The ORF1 sequence contains the gag gene, while genes for an endonuclease (en), reverse transcriptase (rt), and a cysteine-rich domain (Cys) encoding a putative RNA-binding motif are present in ORF2 [12] [23] . The rt is a key enzyme for retrotransposition and shares several conserved domains that are typical of retroviral RNA-directed DNA polymerases [24] . Previously studies have suggested that amplification of rt fragments using degenerate oligonucleotide primers complementary to the conserved domains of the rt is a feasible and efficient approach to evaluate the characterization of LINE retrotransposons in various plant species [18] [25] [26] [27] [28] .
Morus (mulberry) is a representative genus of the cosmopolitan family Moraceae (Rosales), and comprises of more than 13 species (over 1000 cultivars), which are widely distributed in Asia, Africa, Europe, and the United States [29] [30] . Meanwhile, mulberry attracts people for its delicious fruit and rich source of medicines against certain serious diseases [31] [32] . The relationship between mulberry and silkworm is part of the best example of “plant defense-insect adaptation” [33] [34] . The mulberry species, Morus notabilis, has a relatively small genome (estimated to be 357 Mb), and cytogenetic data suggest that M. notabilis is composed of 14 chromosomes (2n = 14) [35] . The previous studies published from our lab are the only papers describing the presence of LINE retrotransposons in the mulberry genome [35] [36] . Detailed characterization of LINE retrotransposable elements in mulberry has not been carried out so far.
In the present work, our objective was to characterize the diversity of rt fragments of LINE retrotransposons from the M. notabilis genome, which were amplified and cloned using degenerate primers. Meanwhile, this present work also attempted to characterize their heterogeneity and phylogenetic relationships. In addition, fluorescence in situ hybridization (FISH) was used to clarify the distribution of these elements within the chromosomes. These results will lead us to better understand the LINE retrotransposons roles on the structural, functional, and evolutionary dynamics of mulberry genomes.
2. Methods
2.1. Plant Materials and DNA Isolation
Young leaves of M. notabilis C.K.Schn (Taxonomy ID: 981085) (2n = 14) were obtained from mulberry trees growing in Ya’an, Sichuan Province, China. The collected young leaves were stored in liquid nitrogen until used. Total genomic DNA used as a template for the cloning was extracted from the young leaves using a standard cetyltrimethylammonium bromide (CTAB) protocol [37] .
2.2. Polymerase Chain Reaction (PCR) and Cloning of Amplicons
The rt sequences of LINEs were amplified from the genomic DNA of mulberry using degenerate primers (forward: 5'-GGGATCCNGGNCCNGAYGGNWT-3'; reverse: 5'-SWNARNGGRTCNCCYTG-3') [18] . The primers were synthesized by BGI (Shenzhen, China). PCR reaction mixture contained 20 ng DNA, 10 pmol of each primer, 0.25 mM of each dNTPs (Takara, Japan), 10× PCR buffer (including 3.5 mM MgCl2, Takara, Japan), and 1 U rTaq polymerase (Takara, Japan). PCR amplification was carried out in 96-well thermal cycler (Applied Biosystems, USA). The PCR program was: 94˚C for 5 min; 35 cycles at 94 °C for 1 min, 50˚C for 1 min, and 72˚C for 1 min; 72˚C for 7 min. PCR products were analyzed on 1.5% agarose gels and purified using the Agarose Gel DNA Extraction Kit (TaKaRa, Japan) according to the manufacturer’s instructions. Purified products were cloned into the pMD19-T vector (TaKaRa, Japan) following the manufacturer’s instructions. Two independent rounds of PCR amplification and cloning were carried out for the elements. The positive clones were verified by PCR and sequenced in both directions using M13 universal primers at Sangon Biotech (Shanghai, China). Clones were named according to the following rules: Mno stands for the Morus notabilis, L means the type of the element (L for LINE), and the serial stands for the clone number from Morus notabilis.
2.3. Sequence Data and Phylogenetic Analysis
Cloned sequences were compared with the previously characterized plant retroelement sequences in the National Center for Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov/) and Genetic Information Research Institute (GIRI) (http://www.girinst.org/) databases using BLAST [38] . The nucleotide and protein sequences were aligned using MUSCLE (version 3.8.31) with default parameters [39] . Sequence identities were calculated by BioEdit (version 7.2.5) with BLOSUM62 matrix [40] . The locations of stop codons in rt sequences were demonstrated using the Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn/) [41] . Nucleotide sequences of the isolated rt fragments and 29 LINE rt sequences from 13 other plant species (Supplementary material 1) were aligned by MUSCLE (version 3.8.31) with default parameters [39] . In order to perform phylogenetic analysis, MEGA6 was used to find the best-fit substitution models for those datasets with default parameters [42] . The best substitution model (Tamura 3-parameter + G, T92 + G) was used to construct a phylogenetic tree according to the maximum-likelihood method with the pairwise deletion in MEGA 6 [42] [43] [44] . Tree topology was assessed by bootstrap analysis with 1000 resampling replicates.
2.4. Chromosome Preparation and FISH
Mulberry chromosome spreads were prepared using young leaves treated with 2 mM 8-hydroxyquinoline in darkness for 3 h at room temperature (24˚C). Samples were fixed in a methanol/glacial acetic acid solution (v/v = 3:1) for 2 h at 4˚C, incubated in 1/15 M KCl for 30 min, and digested by an enzyme mixture (5% cellulose and 2.5% pectinase) at 37˚C for 3 h. After the cell walls were completely degraded, samples were spread onto slides. According to the manufacturer’s instructions for the PCR DIG Probe Synthesis Kit (Roche), probes were labeled with digoxigenin-11-dUTP using PCR with degenerate primers. Fluorescence in situ hybridization was completed according to a modified procedure [45] . Briefly, the prepared chromosomes (on slides) were treated with 100 μg/ml RNase for 15 min at 37˚C, and then digested with 1 μg/ml proteinase K for 10 min at 37˚C. Samples were denatured with 70% (v/v) formamide for 10 min at 72˚C, and then immediately treated for 5 min with each of 70%, 90%, and 100% (v/v) anhydrous ethanol solutions precooled to −20˚C. The hybridization mixture, which consisted of 2× SSC, 0.25 μg salmon sperm DNA, 10% (w/v) SDS, 50% (w/v) DS, 50% (v/v) formamide, and 400 ng labeled DNA probe, was denatured for 6 min at 96˚C. The slides and hybridization mixture were incubated at 80˚C for 10 min and then maintained at 37˚C for 16 h. The slides were washed with 10% (v/v) formamide for 10 s, 2× SSC at 37˚C for 5 min (five times), and 0.2% (v/v) Tween-20 at room temperature for 5 min. Digoxigenin was detected using FITC-conjugated anti-digoxigenin antibody (Roche), and chromosomes were counterstained with 4',6-diamidino-2-phenylindole (DAPI). Slides were viewed using a Leica DM2500 fluorescence microscope (Leica, Germany). Images were captured using the CV-M4+CL progressive scan charge-coupled device camera (DM2500, Leica) and analyzed using CytoVision software (version 7.3.1).
3. Results
3.1. Identification of rt Fragments
The sequences of the expected 580 bp amplicons [18] were compared with sequences available in the NCBI and GIRI databases using BLAST [38] . In total, two independent rounds of PCR and cloning yielded 43 clones with homology to known retroelements in the NCBI and GIRI (http://www.girinst.org/) database. All these elements were selected for further analysis. All the clones are deposited in GenBank under accession numbers: KT900650?KT900692 (Supplementary material 2).
3.2. Characterization of LINE rt Sequences
Nucleotide sequences derived from the isolated rt fragments were aligned and used to construct a phylogenetic tree using the maximum-likelihood method in MEGA6. The identified rt sequences were classified into eight groups (Figure 1). Group I contained the most rt clones (41.8%, 18/43), followed by Group VI (30.2%, 13/43). These two groups accounted for 72.1% of the 43 clones and were further classified into several subfamilies. Additionally, sequences of clones from the same group were of almost the same length, and were highly similar (> 97%, except for Group II) (Table 1 and Supplementary material 2). The length of isolated LINE rt fragments ranged from 557 bp (MnoL_11, MnoL_13, and MnoL_15) to 595 bp (MnoL_31), with an average of 579 bp. The AT/GC ratio
Figure 1. Phylogenetic analysis of reverse transcriptases from mulberry LINE retrotransposons. All cloned nucleotide sequences of reverse transcriptase fragments from mulberry were aligned by muscle (version 3.8.31) under default parameters. The best substitution model (Tamura 3-parameter + G), which was tested by MEGA6, was used to construct a phylogenetic tree based on a maximum-likelihood method with the pairwise deletion in MEGA6. Only more than 50% of the frequency of replicate (1000 replicates) trees were shown. The sequences were classified into eight groups: Group I to Group VIII.
ranged from 1.26 (MnoL_20 and MnoL_37) to 1.98 (MnoL_42), with an average of 1.37, which indicated the rt sequences are rich in AT (Table 1 and Supplementary material 2). Pairwise comparisons revealed that similarity among 43 rt nucleotide sequences ranged from 31.8% (MnoL_20 and MnoL_31, MnoL_31 and MnoL_36) to 99.4% (MnoL_17 and MnoL_29) (Supplementary material 3). These results suggest that the rt sequences isolated were highly heterogeneous.
The alignment of amino acid sequences of multiple isolated mulberry LINE rt rfragments evealed that all sequences contained several premature stop codons, with the exception of the MnoL_10 sequence (Figure 2). The MnoL_42 sequence had 16 premature stop codons, which is the most of any clone. Additionally, the premature stop codons were in similar locations in the sequences from the same group (Figure 2). Furthermore, all sequences carry frameshift mutation, except for the MnoL_10, MnoL_23, and MnoL_31 sequences (Supplementary material 4).
Figure 2. Premature stop codon positions in reverse transcriptase sequences from mulberry LINE retrotransposons. The locations of premature stop codons in rt sequences were demonstrated using the Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn/). Each straight red line represents one of the eight groups of reverse transcriptase sequences (from top to bottom: Group I to Group VIII). The blue line means reverse transcriptase fragments. Red block appeared in the blue line means premature stop codons.
3.3. Phylogenetic Analysis of LINE rt Clones
Phylogenetic analyses indicated that the mulberry LINE rt sequences are homologous to rt sequences in other species (Figure 3). One interesting feature of the result is that PTLRT19 grouped with other mulberry rt sequences (Group V, VI, VII), instead of other PTLRT (PTLRT12, PTLRT4, and PTLRT14) from Paeonia suffruticosa. In fact, P. suffruticosa and M. notabilis are distantly related taxa. The phylogenetic between the LINE-rt sequences and the host species trees were incongruous (APG, The Angiosperm Phylogeny Group, http://www.theplantlist.org).
Figure 3. Phylogenetic analysis of reverse transcriptase fragments from mulberry and other thirteen plant species. All nucleotide sequences of reverse transcriptase fragments from mulberry and the representative members of other thirteen plant species were aligned by muscle (version 3.8.31) with default parameters. Firstly, MEGA6 was used to find the best-fit substitution models for those datasets with default parameters. The best substitution model (Tamura 3-parameter + G) was used to construct a phylogenetic tree, using the maximum-likelihood method with the pairwise deletion in MEGA6. Frequency (>50%) of replicate trees in which the associated taxa clustered together in the bootstraps test (1000 replicates) were shown. Detailed information of other thirteen plant species used in the present research was shown in supplementary material 1.
3.4. Distribution of LINEs in the Mulberry Genome
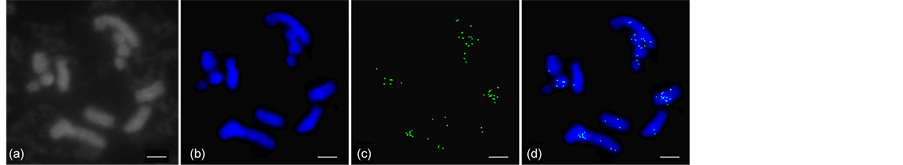
Fluorescence in situ hybridization (FISH) was performed to study the distribution of these sequences along mulberry chromosomes. Chromosomal localization of the LINEs elements was performed using a heterogeneous probe cocktail containing all isolated clones. FISH with such a cocktail revealed that hybridization signals were mainly concentrated in subtelomeric and pericentromeric regions (Figure 4).
4. Discussion
4.1. Characterization of LINEs
All cloned rt fragments could be classified into eight groups (Figure 1). Group I and VI consisted of the most rt clones (72.1%, 31/43). Only one clone (MnoL_42) contained in the Group VIII (Figure 1). The range of nucleotide sequence similarities between MnoL_42 and the other clones was only 32.8 to 37.2% (Supplementary material 3). Meanwhile, the number of premature stop codons in MnoL_42 was 16, which is much higher than in the other clones (Supplementary material 2). These results indicated that mutations accumulated progressively over evolutionary time. Combining these data with the phylogenetic analysis results (Figure 1) allows us to come to the conclusion that the MnoL_42 is an ancient LINE in mulberry.
The rt fragments amplified from the mulberry genome were highly heterogeneous. Almost all the 43 rt sequences described here contained frameshifts and premature stop codons (Figure 2 and Supplementary material 5), which were the main causes of the observed heterogeneity. These results are consistent with those observed in other plants, including Hordeum species [20] and Vicia species [27] . Furthermore, the rt fragments from clones within the same group exhibited very few differences, suggesting that the heterogeneity among rt sequences is also the result of base substitutions, deletions, and insertions. As shown in Figure 2, similar stop codon distribution patterns among sequences of the rt fragments from the same group suggested that they went through a similar evolutionary process.
Figure 4. Chromosomal distribution of mulberry LINE retrotransposons by fluorescence in situ hybridization experiment. Images were captured using the CV-M4+CL progressive scan charge-coupled device camera (DM2500, Leica) and CytoVision software (version 7.3.1). (a) Cytological detection of mulberry chromosome; (b) blue fluorescence signals correspond to 4'-6-diamidino-2-phenylindole-stained DNA; (c) green fluorescence signals correspond to hybridization sites of reverse transcriptase probes; (d) the overlaid images of (b) and (c). Scale bar: 5 μm.
As indicated in Table 1 and Supplementary material 2, the AT/GC ratio ranged from 1.26 to 1.98, with an average of 1.37. The results suggested that the rt sequences are rich in AT bases, which is important for the LINE copy and paste replication mechanism [12] [46] . An intact LINE element contains two open reading frames, ORF1 and ORF2. The ORF2 contains reverse transcriptase, which is a critical enzyme responsible for the replication process of LINE [12] [23] . One of the critical steps in the life cycle of LINE is that the ORF2 protein cleaves the first one DNA strand at the target. Due to the fact that the target sequence in this site is always rich, AT bases and the target site are usually similar to consensus TTAAAA [47] [48] ; the AT bases content is high in the rt sequences to ensure that the target sites can be identified efficiently in the replication process of LINE.
Interestingly, there were no frameshifts or premature stop codons in MnoL_ 10 (Figure 2 and Supplementary material 5). All rt fragments from mulberry, with the exception of clone MonL_10, represented potential pseudogenes (possessed stop codons or frameshifts). It would be worthwhile carrying out further research on MonL_10, which may be a potential active transposable element.
4.2. Phylogenetic Analysis of LINEs
Interestingly, we found a phenomenon that PTLRT19 grouped with other mulberry rt sequences (Group V, VI, and VII), instead of other PTLRT (PTLRT12, PTLRT4, and PTLRT14) from Paeonia suffruticosa (Figure 3). While the two species (P. suffruticosa and M. notabilis) are distantly related taxa according to the APG (The Angiosperm Phylogeny Group, http://www.theplantlist.org). Similarity phenomenon had been found in other studies. For example, twenty-six genomes harbor at least one case of horizontal TE transfer (HTT), which may be important in TE-driven genome evolution, and these HTTs involve species as distantly related as palm and grapevine, tomato and bean, poplar and peach, and so on [49] .
Table 1. Length, AT/GC ratio, and similarity [range (average)] of reverse transcriptase fragments from mulberry LINE retrotransposons.
It is hypothesized that HTT may be the reason for this phenomenon observed in our research. Further analysis was performed in this work. Accordingly, three criteria have been defined for the detection of HTTs: (i) patchy distribution of the TEs in phylogenies, (ii) high sequence similarity of the TE between distantly related taxa, and (iii) phylogenetic incongruence between the TE and host species trees [50] [51] [52] . In the present work, although there is phylogenetic incongruence between the TE and trees of the two species, the range of nucleotide sequence similarities between PTLRT19 and mulberry rt sequences (Group V, VI, and VII) is only 0.520 to 0.562 (Supplementary material 4). These results suggest that the conclusion for PTLRT19 is that it is uncertain whether it represents a horizontal transfer event.
4.3. Chromosomal Localization of LINE Retrotransposons
The LINE distribution patterns are associated with LINE functions. For example, FISH experiments in Cannabis sativa suggested that differential accumulation of LINE retrotransposon elements onto the Y chromosome leads to sex chromosome heteromorphism [53] . Although the chromosomal distribution of LINEs has been analyzed in only a few plant species, the FISH results here revealed that the distribution of LINEs in mulberry chromosomes was similar to that in sugar beet and peanut chromosomes [28] [54] . Furthermore, the weak hybridization signals observed in this study indicated that LINEs were not abundant in the mulberry genome (Figure 4). Most of the hybridization signals were concentrated in subtelomeric and pericentromeric regions. The subtelomeric and pericentromeric regions are generally considered to correspond to the constitutively heterochromatic region. In fact, there is extensive DNA methylation in these regions [55] . Thus, we hypothesized that the tendency for LINEs to insert into these regions may be related to DNA methylation in mulberry.
Meanwhile, recent reports suggest that LINE insertion into promoters can influence promoter functionality and gene regulation, resulting in up or down regulation of reporter genes [56] . Insertion of a LINE into a gene can induce alternative splicing or change gene expression patterns, which can result in a change in the function of the gene [57] . Although we currently have no evidence that the mulberry LINEs described here are active and functional, previous studies have indicated that some LINEs are active and functional in other species [58] . So, our future studies will attempt to characterize the functions of mulberry LINEs more comprehensively, considering their localization in subtelomeric and pericentromeric regions.
Acknowledgements
The authors declare that they have no conflict of interest. The authors thank all the laboratory members who provided advice during this work. This project was funded by “Fundamental Research Funds for the Central Universities (XDJK2015C116)” and “China Postdoctoral Science Foundation (2016M592622)”.
Conflict of Interest
All authors declare that there is no conflict of interest.
Cite this paper
Ma, B., Xin, Y.C., Kuang, L.L., Hou, F. and He, N.J. (2017) Identification and Characterization of Re- verse Transcriptase Fragments of Long In- terspersed Nuclear Elements (LINEs) in the Morus notabilis Genome. American Journal of Molecular Biology, 7, 138-152. https://doi.org/10.4236/ajmb.2017.73011
References
- 1. McClintock, B. (1950) The Origin and Behavior of Mutable Loci in Maize. Proceedings of the National Academy of Sciences of the United States of America, 36, 344-355.
https://doi.org/10.1073/pnas.36.6.344 - 2. Bucher, E., Reinders, J. and Mirouze, M. (2012) Epigenetic Control of Transposon Transcription and Mobility in Arabidopsis. Current Opinion in Plant Biology, 15, 503-510.
https://doi.org/10.1016/j.pbi.2012.08.006 - 3. Bennetzen, J.L. (2000) Transposable Element Contributions to Plant Gene and Genome Evolution. Plant Molecular Biology, 42, 251-269.
https://doi.org/10.1023/A:1006344508454 - 4. Feschotte, C. (2008) Transposable Elements and the Evolution of Regulatory Networks. Nature Reviews Genetics, 9, 397-405.
https://doi.org/10.1038/nrg2337 - 5. Mita, P. and Boeke, J.D. (2016) How Retrotransposons Shape Genome Regulation. Current Opinion in Genetics & Development, 37, 90-100.
https://doi.org/10.1016/j.gde.2016.01.001 - 6. Lisch, D. (2013) How Important Are Transposons for Plant Evolution? Nature Reviews Genetics, 14, 49-61.
https://doi.org/10.1038/nrg3374 - 7. Rajput, M.K. (2015) Retrotransposons: The Intrinsic Genomic Evolutionist. Genes & Genomics, 37, 113-123.
https://doi.org/10.1007/s13258-014-0250-5 - 8. Kumar, A. and Bennetzen, J.L. (1999) Plant Retrotransposons. Annual Review of Genetics, 33, 479-532.
https://doi.org/10.1146/annurev.genet.33.1.479 - 9. Du, J., Tian, Z., Hans, C.S., Laten, H.M., Cannon, S.B., Jackson, S.A., Shoemaker, R.C. and Ma, J. (2010) Evolutionary Conservation, Diversity and Specificity of LTR-Retrotransposons in Flowering Plants: Insights from Genome-Wide Analysis and Multi-Specific Comparison. The Plant Journal, 63, 584-598.
https://doi.org/10.1111/j.1365-313X.2010.04263.x - 10. Wicker, T., Sabot, F., Hua-Van, A., Bennetzen, J.L., Capy, P., Chalhoub, B., Flavell, A., Leroy, P., Morgante, M., Panaud, O., et al. (2007) A unified Classification System for Eukaryotic Transposable Elements. Nature Reviews Genetics, 8, 973-982.
https://doi.org/10.1038/nrg2165 - 11. Noma, K., Ohtsubo, E. and Ohtsubo, H. (1999) Non-LTR Retrotransposons (LINEs) as Ubiquitous Components of Plant Genomes. Molecular and General Genetics, 261, 71-79.
https://doi.org/10.1007/s004380050943 - 12. Schmidt, T. (1999) LINEs, SINEs and Repetitive DNA: Non-LTR Retrotransposons in Plant Genomes. Plant Molecular Biology, 40, 903-910.
https://doi.org/10.1023/A:1006212929794 - 13. Heitkam, T. and Schmidt, T. (2009) BNR—A LINE Family from Beta vulgaris—Contains a RRM Domain in Open Reading Frame 1 and Defines a L1 Sub-Clade Present in Diverse Plant Genomes. The Plant Journal, 59, 872-882.
https://doi.org/10.1111/j.1365-313X.2009.03923.x - 14. Belancio, V.P., Hedges, D.J. and Deininger, P. (2008) Mammalian Non-LTR Retrotransposons: For Better or Worse, in Sickness and in Health. Genome Research, 18, 343-358.
https://doi.org/10.1101/gr.5558208 - 15. Han, J.S. (2010) Non-Long Terminal Repeat (Non-LTR) Retrotransposons: Mechanisms, Recent Developments, and Unanswered Questions. Mobile DNA, 1, 15.
https://doi.org/10.1186/1759-8753-1-15 - 16. Schwarzsommer, Z., Leclercq, L., Gobel, E. and Saedler, H. (1987) Cin4, an Insert Altering the Structure of the A1 Gene in Zea mays, Exhibits Properties of Nonviral Retrotransposons. The EMBO Journal, 6, 3873-3880.
- 17. Leeton, P.R. and Smyth, D.R. (1993) An Abundant LINE-Like Element Amplified in the Genome of Lilium speciosum. Molecular and General Genetics, 237, 97-104.
https://doi.org/10.1007/bf00282789 - 18. Wright, D.A., Ke, N., Smalle, J., Hauge, B.M., Goodman, H.M. and Voytas, D.F. (1996) Multiple Non-LTR Retrotransposons in the Genome of Arabidopsis thaliana. Genetics, 142, 569-578.
- 19. Higashiyama, T., Noutoshi, Y., Fujie, M. and Yamada, T. (1997) Zepp, a LINE-Like Retrotransposon Accumulated in the Chlorella Telomeric Region. The EMBO Journal, 16, 3715-3723.
https://doi.org/10.1093/emboj/16.12.3715 - 20. Vershinin, A.V., Druka, A., Alkhimova, A.G., Kleinhofs, A. and Heslop-Harrison, J.S. (2002) LINEs and gypsy-Like Retrotransposons in Hordeum Species. Plant Molecular Biology, 49, 1-14.
https://doi.org/10.1023/A:1014469830680 - 21. Komatsu, M., Shimamoto, K. and Kyozuka, J. (2003) Two-Step Regulation and Continuous Retrotransposition of the Rice LINE-Type Retrotransposon Karma. The Plant Cell, 15, 1934-1944.
https://doi.org/10.1105/tpc.011809 - 22. Yamashita, H. and Tahara, M. (2006) A LINE-Type Retrotransposon Active in Meristem Stem Cells Causes Heritable Transpositions in the Sweet Potato Genome. Plant Molecular Biology, 61, 79-84.
https://doi.org/10.1007/s11103-005-6002-9 - 23. Piskareva, O., Ernst, C., Higgins, N. and Schmatchenko, V. (2013) The Carboxy-Terminal Segment of the Human LINE-1 ORF2 Protein Is Involved in RNA Binding. FEBS Open Bio, 3, 433-437.
https://doi.org/10.1016/j.fob.2013.09.005 - 24. Xiong, Y. and Eickbush, T.H. (1990) Origin and Evolution of Retroelements Based upon Their Reverse Transcriptase Sequences. The EMBO Journal, 9, 3353-3362.
- 25. Malik, H.S., Burke, W.D. and Eickbush, T.H. (1999) The Age and Evolution of Non-LTR Retrotransposable Elements. Molecular Biology and Evolution, 16, 793-805.
https://doi.org/10.1093/oxfordjournals.molbev.a026164 - 26. Kubis, S.E., Castilho, A.M., Vershinin, A.V. and Heslop-Harrison, J.S. (2003) Retroelements, Transposons and Methylation Status in the Genome of Oil Palm (Elaeis guineensis) and the Relationship to Somaclonal Variation. Plant Molecular Biology, 52, 69-79.
https://doi.org/10.1023/A:1023942309092 - 27. Hill, P., Burford, D., Martin, D.M. and Flavell, A.J. (2005) Retrotransposon Populations of Vicia Species with Varying Genome Size. Molecular Genetics and Genomics, 273, 371-381.
https://doi.org/10.1007/s00438-005-1141-x - 28. Samoluk, S.S., Robledo, G., Podio, M., Chalup, L., Ortiz, J.P.A., Pessino, S.C. and Seijo, J.G. (2015) First Insight into Divergence, Representation and Chromosome Distribution of Reverse Transcriptase Fragments from L1 Retrotransposons in Peanut and Wild Relative Species. Genetica, 143, 113-125.
https://doi.org/10.1007/s10709-015-9820-y - 29. Nepal, M.P. and Ferguson, C.J. (2012) Phylogenetics of Morus (Moraceae) Inferred from ITS and trnL-trnF Sequence Data. Systematic Botany, 37, 442-450.
https://doi.org/10.1600/036364412X635485 - 30. Berg, C.C. (2005) Moraceae Diversity in a Global Perspective. Proceedings of a Symposium on Plant Diversity and Complexity Patterns—Local, Regional and Global Dimensions. Biologiske Skrifter, 55, 423-440.
- 31. Venkatesh, K.P. and Chauhan, S. (2008) Mulberry: Life Enhancer. Journal of Medicinal Plants Research, 2, 271-278.
- 32. Darias-Martin, J., Lobo-Rodrigo, G., Hernandez-Cordero, J., Diaz-Diaz, E. and Diaz-Romero, C. (2003) Alcoholic Beverages Obtained from Black Mulberry. Food Technology and Biotechnology, 41, 173-176.
- 33. Konno, K., Ono, H., Nakamura, M., Tateishi, K., Hirayama, C., Tamura, Y., Hattori, M., Koyama, A. and Kohno, K. (2006) Mulberry Latex Rich in Antidiabetic Sugar-Mimic Alkaloids Forces Dieting on Caterpillars. Proceedings of the National Academy of the United States of America, 103, 1337-1341.
https://doi.org/10.1073/pnas.0506944103 - 34. Daimon, T., Taguchi, T., Meng, Y., Katsuma, S., Mita, K. and Shimada, T. (2008) Beta-Fructofuranosidase Genes of the Silkworm, Bombyx mori: Insights into Enzymatic Adaptation of B. mori to Toxic Alkaloids in Mulberry Latex. Journal of Biological Chemistry, 283, 15271-15279.
https://doi.org/10.1074/jbc.M709350200 - 35. He, N., Zhang, C., Qi, X., Zhao, S., Tao, Y., Yang, G., Lee, T.H., Wang, X., Cai, Q., Li, D., et al. (2013) Draft Genome Sequence of the Mulberry Tree Morus notabilis. Nature Communications, 4, Article No. 2445.
https://doi.org/10.1038/ncomms3445 - 36. Ma, B., Li, T., Xiang, Z. and He, N. (2015) MnTEdb, a Collective Resource for Mulberry Transposable Elements. Database (Oxford), 2015, bav004-bav004.
https://doi.org/10.1093/database/bav004 - 37. Doyle, J. (1991) DNA Protocols for Plants. In: Hewitt, G.M., Johnston, A.W.B. and Young, J.P.W., Eds., Molecular Techniques in Taxonomy, Springer, Berlin, Heidelberg, 283-293.
https://doi.org/10.1007/978-3-642-83962-7_18 - 38. Altschul, S.F., Gish, W., Miller, W., Myers, E.W. and Lipman, D.J. (1990) Basic Local Alignment Search Tool. Journal of Molecular Biology, 215, 403-410.
- 39. Edgar, R.C. (2004) MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Research, 32, 1792-1797.
https://doi.org/10.1093/nar/gkh340 - 40. Hall, T. (2011) BioEdit: An Important Software for Molecular Biology. GERF Bulletin of Bioscience, 2, 6.
- 41. Guo, A.Y., Zhu, Q.H., Chen, X. and Luo, J.C. (2007) [GSDS: A Gene Structure Display Server]. Hereditas, 29, 1023-1026.
- 42. Tamura, K., Stecher, G., Peterson, D., Filipski, A. and Kumar, S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution, 30, 2725-2729.
https://doi.org/10.1093/molbev/mst197 - 43. Yang, Z. (1996) Maximum-Likelihood Models for Combined Analyses of Multiple Sequence Data. Journal of Molecular Evolution, 42, 587-596.
https://doi.org/10.1007/BF02352289 - 44. Tamura, K. (1992) Estimation of the Number of Nucleotide Substitutions When There Are Strong Transition-Transversion and G+C-Content Biases. Molecular Biology and Evolution, 9, 678-687.
- 45. Schwarzacher, T. and Heslopharrison, P. (2000) Practical in Situ Hybridization. Heredity, 85, 97-97.
- 46. Dhellin, O., Maestre, J. and Heidmann, T. (1997) Functional Differences between the Human LINE Retrotransposon and Retroviral Reverse Transcriptases for in Vivo mRNA Reverse Transcription. The EMBO Journal, 16, 6590-6602.
https://doi.org/10.1093/emboj/16.21.6590 - 47. Kaessmann, H., Vinckenbosch, N. and Long, M. (2009) RNA-Based Gene Duplication: Mechanistic and Evolutionary Insights. Nature Reviews Genetics, 10, 19-31.
https://doi.org/10.1038/nrg2487 - 48. Rawal, K. and Ramaswamy, R. (2011) Genome-Wide Analysis of Mobile Genetic Element Insertion Sites. Nucleic Acids Research, 39, 6864-6878.
https://doi.org/10.1093/nar/gkr337 - 49. El Baidouri, M., Carpentier, M.C., Cooke, R., Gao, D., Lasserre, E., Llauro, C., Mirouze, M., Picault, N., Jackson, S.A. and Panaud, O. (2014) Widespread and Frequent Horizontal Transfers of Transposable Elements in Plants. Genome Research, 24, 831-838.
https://doi.org/10.1101/gr.164400.113 - 50. Schaack, S., Gilbert, C. and Feschotte, C. (2010) Promiscuous DNA: Horizontal Transfer of Transposable Elements and Why It Matters for Eukaryotic Evolution. Trends in Ecology & Evolution, 25, 537-546.
https://doi.org/10.1016/j.tree.2010.06.001 - 51. Silva, J.C., Loreto, E.L. and Clark, J.B. (2004) Factors That Affect the Horizontal Transfer of Transposable Elements. Current Issues in Molecular Biology, 6, 57-71.
- 52. Loreto, E.L., Carareto, C.M. and Capy, P. (2008) Revisiting Horizontal Transfer of Transposable Elements in Drosophila. Heredity, 100, 545-554.
https://doi.org/10.1038/sj.hdy.6801094 - 53. Sakamoto, K., Ohmido, N., Fukui, K., Kamada, H. and Satoh, S. (2000) Site-Specific Accumulation of a LINE-Like Retrotransposon in a Sex Chromosome of the Dioecious Plant Cannabis sativa. Plant Molecular Biology, 44, 723-732.
https://doi.org/10.1023/A:1026574405717 - 54. Kubis, S.E., Heslop-Harrison, J.S., Desel, C. and Schmidt, T. (1998) The Genomic Organization of Non-LTR Retrotransposons (LINEs) from Three Beta Species and Five Other Angiosperms. Plant Molecular Biology, 36, 821-831.
https://doi.org/10.1023/A:1005973932556 - 55. Suzuki, M.M. and Bird, A. (2008) DNA Methylation Landscapes: Provocative Insights from Epigenomics. Nature Reviews Genetics, 9, 465-476.
https://doi.org/10.1038/nrg2341 - 56. Dhadi, S.R., Xu, Z., Shaik, R., Driscoll, K. and Ramakrishna, W. (2015) Differential Regulation of Genes by Retrotransposons in Rice Promoters. Plant Molecular Biology, 87, 603-613.
https://doi.org/10.1007/s11103-015-0300-7 - 57. Xu, Z.J. and Ramakrishna, W. (2008) Retrotransposon Insertion Polymorphisms in Six Rice Genes and Their Evolutionary History. Gene, 412, 50-58.
- 58. Hu, G., Hawkins, J.S., Grover, C.E. and Wendel, J.F. (2010) The History and Disposition of Transposable Elements in Polyploid Gossypium. Genome, 53, 599-607.
https://doi.org/10.1139/G10-038
All Links to Supplementary Materials Were Created and Listed Below
Supplementary Material 1:
https://drive.google.com/open?id=0BwZN0JGYCBPNbmhweU1QT01nZkE
Supplementary Material 2:
https://drive.google.com/open?id=0BwZN0JGYCBPNSVhhdkNXU2YzdUE
Supplementary Material 3:
https://drive.google.com/open?id=0BwZN0JGYCBPNVjdXd1lPaURyNUU
Supplementary Material 4:
https://drive.google.com/open?id=0BwZN0JGYCBPNQVllSmQ1MUhIWlk
Supplementary Material 5
https://drive.google.com/open?id=0BwZN0JGYCBPNVTBjaXVmSm9UMzg

Submit or recommend next manuscript to SCIRP and we will provide best service for you:
Accepting pre-submission inquiries through Email, Facebook, LinkedIn, Twitter, etc.
A wide selection of journals (inclusive of 9 subjects, more than 200 journals)
Providing 24-hour high-quality service
User-friendly online submission system
Fair and swift peer-review system
Efficient typesetting and proofreading procedure
Display of the result of downloads and visits, as well as the number of cited articles
Maximum dissemination of your research work
Submit your manuscript at: http://papersubmission.scirp.org/
Or contact ajmb@scirp.org
NOTES
*These authors contribute equally to this work.