American Journal of Plant Sciences
Vol.3 No.6(2012), Article ID:20051,6 pages DOI:10.4236/ajps.2012.36098
Identification of DNA Photolyase Gene of a Drought Tolerance Branching Line of Kenaf (Hibiscus cannabinus L.)
![]()
1Laboratory of Molecular Biology, Department of Biology, Faculty of Mathematics and Natural Sciences, Brawijaya University, Malang, Indonesia; 2Laboratory of Plant Breeding, Faculty of Agriculture, Brawijaya University, Malang, Indonesia; 3Research Institute for Tobacco and Fibre Crops, Malang, Indonesia.
Email: {laras, sutiman}@ub.ac.id, sudjindro@telkom.net
Received April 18th, 2012; revised May 14th, 2012; accepted May 24th, 2012
Keywords: DNA Photolyase; Kenaf; Detection
ABSTRACT
DNA photolyase is a photoreactivation enzyme which has a role in the mechanism of DNA repair. The existence of DNA photolyase gene could affect the success of mutation process. SM026H is a drought tolerance branching line of kenaf identified by Research Institute for Tobacco and Fibre Crops, Karangploso, Malang, Indonesia which has a potential as a source for mutation breeding. In this research, cloning and sequencing of the DNA photolyase gene of line SM026H were conducted. The DNA of SM026H leaves was isolated using DNeasy Kit, then was amplified by Polymerase Chain Reaction (PCR) using AC1 as a forward primer, paired with AC3R or AC4R as reverse primers and checked using electrophoresis on a 0.7% agarose gel. The AC1-AC3R primer pair produced a DNA fragment with a size of 750 bp, whereas the AC1-AC4R primer pair produced a DNA fragment of 1000 bp in size. Cloning of the AC1-AC3R and AC1-AC4R fragments was done prior to sequencing. Preparation for cloning was done by running and extracting the PCR products from a 0.7% Low Melting Agarose (LMA), and then ligating them to a pCR21 plasmid using an electroporation method. Two sub-fragments of each AC1-AC3R and AC1-AC4R fragments were identified. They were 600 bp and 500 bp, resulting from the AC1-AC3R fragment, and 1300 bp and 500 bp, resulting from the AC1-AC4R fragment. The sequencing result of those sub-fragments was analyzed using a Basic Local Alignment Search Tool (BLAST) program. It was shown that the sequences have a degree of homology to DNA photolyase sequence of Arabidopsis thaliana A. thaliana, Cucumis sativus, Spinacia oleracea, Stellaria longipes and Oryza sativa. These suggested that the kenaf line SM0026H possesses the DNA photolyase gene. However, further study needs to be done to ascertain that the gene was not the cryptochrome.
1. Introduction
Kenaf (Hibiscus cannabinus L.) is a fiber plant which belongs to the Malvaceae family whose bast fiber is usually utilized for making ropes, sacks, canvases and carpets. Kenaf usage has been developed for wider purposes, including for pulping and paper making, oil absorption, potting media, board making, filtration media and animal feed [1-3]. Many research studies have been conducted to maximize the utilization of kenaf. Research projects for producing cultivated varieties which increased the biomass of the kenaf plant were developed to fulfil the global demand for a sufficient and secure supply of raw materials for paper pulp [4]. The kenaf line SM026H is a drought-tolerance line which is one of germplasm collection of the Research Institute for Tobacco and Fibre Crops, Karangploso, Malang, Indonesia [5]. This line has a short stem and exhibits many branches. The gene that converts branching phenotypes is very important for the source of DNA which is used in the induction of higher biomass lines.
One method of producing a new variety in plant breeding is radiation mutation. Ultra-violet UV radiation has been widely known capable to induce mutation on plants [6-9]. Despite its advantages for inducing mutation, (UV) radiation in the sunlight stimulates lesions to form in DNA. When DNA is damaged by UV, 75% is by the formation of cyclobutan pyrimidin dimer (CPD) and 25% is by the formulation of 6-4 photoproduct (6-4 PPs) [10]. The UV-induced lesions in DNA block the replication and transcription events in the living cells, causing growth delay, mutagenesis, or lethal effects on organisms [6,11, 12] and decreasing productivity of plants [6,11]. Higher plants repair the DNA damage caused by UV mainly by photoreactivation [7,10,13-15], utilizing light-dependent DNA-repair enzymes called DNA photolyases. These enzymes use blue or near-UV light (350 - 450 nm) as an energy source [12,16-19]. CPDs are removed by a photolyase photo-reactivating enzyme 1/UV resistant 2(PHR1/ UVR2), which structurally contains 2 chromophores, flavin FAD cofactors which act as electron donors which revert CPDs in free error manner [20,21] and other chromophores that confer specificity [22]. 6-4 PPs are removed by another photolyase gene, UVR3 [23]. The CPDs, 6-4 PPs and cryptochromes (Crys) are three major groups of protein constituting a Cryptochrome/photolyase family (CPF). All the members of this family are flavoproteins that use near-ultraviolet (UV)/blue light photons as an energy source to mediate either DNA repairs or perceptions and signal transduction of external light signals. Members of this superfamily are widely distributed throughout bacteria, archaea and eukaryotes [18]. Whilst CPDs and 6-4 PPs act as agents for repairing DNA damage, Cryptochromes actas blue-light photoreceptors and exert various physiological functions, such as regulation of the circadian clock in animals and plants. Unlike photolyases, they generally lack of any kind of DNA-repair activity [18].
Research on CPD photolyases has been conducted using various organisms including Anacytis nidulans, Arabidopsis thaliana, Chlamydomonas, Glycine max and Oryza sativa. It has been detected that the DNA sequence of this enzyme on Clamydomonas and Arabidopsis thaliana are highly homolog. So, two pairs of primers were designed based on the CPD photolyase sequence of the two organisms, and were used to detect the DNA photolyase gene of the kenaf line SM026H.
2. Material and Methods
2.1. DNA Isolation and PCR for the DNA Photolyase Gene of Kenaf
The DNA genome was isolated from young leaves of kenaf SM026H using Dneasy Kit (Qiagen). A forward primer AC1 and two reverse primers, AC3R and AC4R, which were designed based on the DNA sequences of Clamydomonas and Arabidopsis thaliana photolyase enzyme, were used to prime the DNA photolyase sequence of kenaf. These primers were mixed with 5 ul 10× buffer Taq Polymerase, 4 ul dNTP, 4 ul 2mM MgCl2 and 0.5 ul Ex Taq Polymerase and used for PCR reaction.
The condition used was 40 cycles of PCR reaction consisting of 1 minute denaturation at the temperature of 93˚C, 1 minute annealing at the temperature of 50˚C, and 1 minute and 30 seconds extension at the temperature of 72˚C. A preheating period was applied for 1 minute at the temperature of 93˚C, and the last elongation step was conducted at 72˚C for 10 minutes.
2.2. Electrophoresis DNA, Ligation, and Transformation
An electrophoresis on 0.7% agarose with a TAE buffer was conducted to check the DNA which resulted from the PCR process. When the expected band was confirmed, electrophoresis using Low Melting Agarose (LMA) 0.7% was performed. The resulting bands were extracted from LMA using a phenol solution and then were precipitated using an ethanol solution. The DNA extracted from the LMA was ligated to 1 ul vector, pCR 21, using a 30 ul reaction mixture consisting of 3 ul 10× buffer T4 ligase and ddH2O and incubated at 16˚C overnight. The DNA donor and the plasmid were cut using EcoRV restriction enzyme. The ligation result then was reprecipitated using ethanol and transformed to E. coli strain XL-1 Blue, using the electroporation method. The transformed bacteria were spread on a selection medium containing ampicillin for the white blue selection. The succeeding transformed cells (white colony) were taken and streaked on a new medium, and incubated for 10 hours. The white colony was then taken into liquid LB containing ampicillin medium and incubated at 37˚C overnight.
2.3. Plasmid Extraction, Restriction Checking and Double Strand Sequencing
Plasmid which had been inserted with the DNA sequence of the DNA photolyase gene was extracted from E. coli and checked by cutting the plasmid with a restriction enzyme. The right plasmid then was treated with Rnase, extracted with phenol and continued with Polyethylene Glycol extraction (13% PEG 65 µl + 5M NaCl 15 µl + 50 µl sample = 130 µl). Prior to the sequencing procedure, PCR was conducted using a 20 µl solution mixture of 200 - 500 ng DNA, 0.5 µl primer, 8 µl Dyenamic ET term. Seq. kit and ddH2O. The condition used consisted of 1 minute preheating at 95˚C, continued with 25 cycles of denaturation for 20 seconds at 95˚C, annealing for 15 seconds at 50˚C and extention for1 minute at 60˚C. The results were precipitated using ethanol and finally were dissolved in 20 µl Template Supression Reagent (TSR), and put into a sequencing machine.
3. Result
Research on the molecular characteristic of kenaf is limited. There is no information about kenaf genome sequence. In the absence of kenaf genome sequence information, it was decided to utilize primers which were formerly used for cloning photolyase genes in soybeans which were designed based on the conserved region of the amino acid motif of Class II DNA photolyases of Arabidopsis thaliana and Chlamydomonas reinhardtii [24]. Using the degenerate primers AC1-AC3R, and AC1-AC4R,PCR fragments with the sizes of approximately 750 bp and 1000 bp, respectively, were developed (Figure 1).
Each PCR fragment then was cloned and sequenced to detect the relative position of the fragments on the photolyase gene. Confirmation of the effectiveness of

Figure 1. PCR result using primers AC1-AC3R and AC1- AC4R.
transformation and detection of sub-fragments of the PCR product, which was done using the restriction digestion method, was capable of identifying two sub-fragments (0.6 kb and 0.5 kb) derived from AC1-AC3R and two sub-fragments (1.3 kb and 0.5 kb) derived from AC1-AC4R (Figure 2). Apparently, the PCR products which seem to be single bands consist of more than one fragment.
Using the Basic Local Alignment Search Tool (BLAST) system, it was detected that the sequence of the sub-fragments has some degree of homology with the mRNA for type II CPD photolyase, PHR1, complete cds of some plant species, A. thaliana, Cucumis sativus, Spinacia oleracea, Stellaria longipes and Oryza sativa (Table 1). However, there was no evidence of 100% homology, showing the existence of sequence variation of kenaf’s CPD photolyase compared to those of other plant species.
The first sequence AC1-AC3R (500 bp), has only a short sequence homology with A. thaliana class II CPD photolyase. The number of nitrogen bases which were similar to the A. thaliana class II CPD photolyase was about 40 bases out of 367 bases examined. A similar result was also shown by the second sequence (AC1-AC3R 2), and the sequence of AC1-AC4R (Table 1). They had only 90 to 140 bases of homology out of 428 bp and 50 out of 400 bases, respectively, to other CPD photolyases of some plants which has been compared.
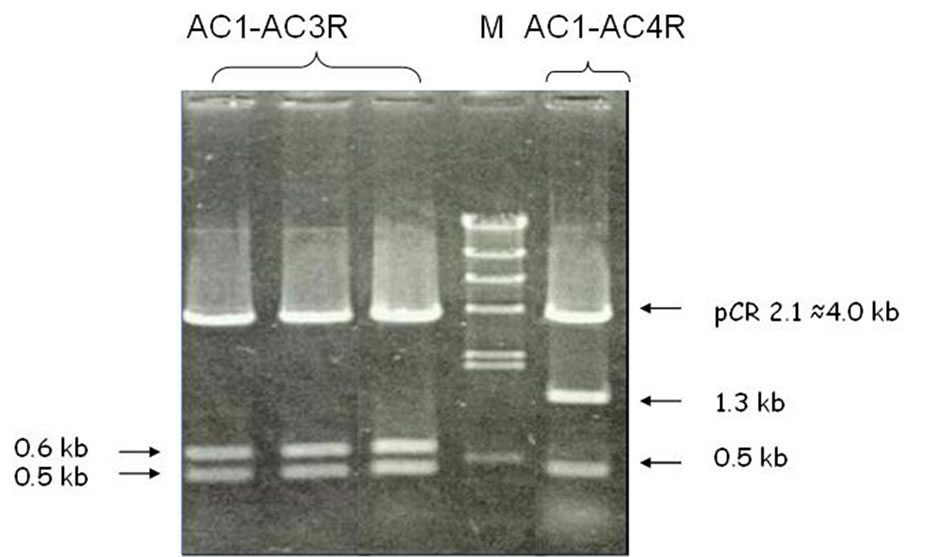
Figure 2. Restriction digestion of recombinant plasmid showing sub-fragments of AC1-AC3R and AC1-AC4R PCR fragment.

Table 1. Sequence homology of fragments AC1-AC3R (0.5 Kb), AC1-AC4R (0.6 Kb), AC1-AC4R (0.5 Kb) with DNA Photolyase of other organisms.
By comparing the length of sub-fragments which resulted from the cloning, and the sequence of those fragments, the order of those sub-fragments was predicted and presented in Figure 3.
The 500 bp sub-fragment of AC1-AC3R has a similar size and sequence to the 500 bp sub-fragment of AC1- AC4R. The second sub-fragment of AC1-AC3R, with the size of 600 bp, was overlapped with part of the sub-fragment 1300 bp of the AC1-AC4R fragment. Considering the direction of primers used and the sequence of all subfragments, the 500 bp sub-fragment was positioned downward to the other sub-fragments.
4. Discussion
The primers used in this experiment were capable of identifying the existence of the CPD photolyase gene in kenaf. However, the sequence of the gene was not 100% similar to other CPD photolyases already identified in other plants such as Arabidopsis thaliana, Cucumis sativus, Spinacia oleracea, Stellaria longipes and Oryza sativa. The homology analysis using BLAST suggests that the sequences identified in this experiment is a type II CPD photolyase, which is common in eukaryotes [17,25,26]. However, the partial similarity revealed by those sequences showed that the kenaf CPD photolyase is quite different from the other photolyases identified. Those differences may be evidence that the CPD photolyase gene of kenaf, indeed, has a different sequence from other CPD photolyases, but it is positioned in the same locus and still has the ability to repair DNA photolessions. Another possibility is that the identified sequence is a different gene in a different locus which has some sequence similarity to CPD photolyase. This would mean that this sequence has no repair ability. So, those differences need to be further investigated, since they have a significant effect on the breeding strategy of kenaf. The line SM026H is a derivative of a wild type kenaf. This line has a short stem and profuse branches and has the potential of resistance to nematodes which is the main disease of kenaf. However, it is preferred that commercial lines of kenaf have long stems and no branches. Breeding kenaf commonly was done by crossing between lines, or by mutagenesis [4], among which is UV mutagenesis. The presence of a photolyase gene will prevent the success of a breeding program using the UV mutagenesis method. On the other hand, to maintain the mutants stability under natural UV when they are grown in the field

Figure 3. Predicted position of fragments in the kenaf photolyase gene.
requires the existence of a CPD gene. So, it is important to determine the kind of gene possessed by the kenaf SM26 line.
The CPD photolyase is a member of the superfamily cryptochrome/photolyase family, together with 6-4 PPs and Cryptochromes (Crys). Both photolyases and cryptochromes have a barbell-like structure consisting of two cofactors: the first cofactor is a C-terminal helical catalytic domain comprising the light-sensitive flavin adenine dinucleotide (FAD) cofactor [17] and the second cofactor is an N-terminal domain that may contain a light-harvesting antenna, chromophore, to additionally broaden their activity spectra in order to increase the repair efficiency of the enzymes under limited light conditions. This N-terminal domain usually is a derivative of folate, deazaflavin or flavin [17,27,28].
Considering the differences found in the sequence of kenaf CPD can lead to the rational that the sequence could be a cryptochrome gene. The possibility needs to be taken into account, since there has been no information about that available yet. It is known that most of the plant cryptochrome has a size of 70 - 80 kD with two domains, namely the amino end domain of PHR (photolyase related) that has a homology with photolyase and carboxyl ends, which are typical for cryptochromes. On the other hand, photolyase are 55 - 65 kD in size, containing two chromophores, namely methenyltetrahydrofolate deazaflavin and flavin adenine dinucleotide (FAD) (Lin, 2002). Almost all residues that are essential for binding of chromophores in photolyase are homologous with the residues in cryptochrome, but the residues that play a role in the binding and repair of DNA lesions are not owned by the cryptochrome [29]. To ensure whether the sequence identified in this research is the cryptochrome, further investigation using isoenzym analysis needs to be conducted, referring to the differences in the size of CPD photolyase and cryptochrome.
Furthermore, to prove the real activity of this enzyme on kenaf, research needs to be done in order to detect the effect of UV light on the mutagenesis of CPD photolyase-contained kenaf. Among others is by detecting and comparing the expression level of photolyase gene in kenaf mutant and the initial line. Information about this gene will greatly contribute to the strategy of breeding a new variety of kenaf, such as a UV resistant-kenaf line.
5. Conclusion
The ability of the CPD primers used in this experiment to amplify and produce DNA fragments, which have some degree of homology to CPD photolyase of some reference plants, indicates that kenaf possesses the gene. However, since the homology of the sequence to the CPD photolyase already recorded was quite low, further investigation needs to be conducted. Identification of protein products which is expressed by the CPD photolyase gene and its ability of repairing DNA photolessions need to be done.
6. Acknowledgements
We are grateful to the Research Institute for Tobacco and Fibre Crops, Karangploso, Malang, Indonesia, for providing kenaf seeds, Tokuhisha Hirouchi for technical assistance, Prof. Kazuo Yamamoto for providing research facilities, and Dr. Larry Cothren from the Intensive English Language Institute (IELI) Flinders University, South Australia, for English language assistance with the manuscript. The research was supported by the Directorate General of Higher Education (DGHE)’s PHB project, Indonesia.
REFERENCES
- C. S. Taylor, “Kenaf,” Kenaf International, Ltd., McAllen, 1995. http://www.hort.purdue.edu/newcrop/CropFactSheets/kenaf.html
- C. L. Webber III, J. Whitworth and J. Dole, “Kenaf (Hibiscus cannabinus L.) Core as a Containerized Growth Medium Component,” Industrial Crops and Products, Vol. 10, No. 2, 1999, pp. 97-105. doi:10.1016/S0926-6690(99)00014-X
- A. Tsakonas, V. Stergiou and M. Polissiou, “Kenaf (Hibiscus cannabinus L.) Based Substrates for the Production of Compact Plants,” Industrial Crops and Products Vol. 21, No. 2, 2005, pp. 223-227. doi:10.1016/j.indcrop.2004.04.008
- E. L. Arumingtyas, A. Munawarti, S. Indriyani and Sudjindro, “Polymorphism Analysis of Kenaf (Hibiscus Cannabinus L.) Mutants Based on Random Amplified Polymorphic DNAs (RAPDs),” Journal of Materials Science and Engineering, Vol. 4, No. 2, 2010, pp. 56-62.
- U. Setyobudi, R. D. Purwati, D. I. Kangiden and S. Hartati, “Asesment of Some Kenaf Lines against Drought Stress,” Research Institute for Tobacco and Fibre Crops Malang, Vol. 8, No. 2, 1993, pp. 104-111.
- B. R. Jordan, “The Effects of Ultraviolet-B Radiation on Plants: A Molecular Perspective,” In: J. A. Callow, Ed., Advances in Botanical Research Incorporating Advances in Plant Pathology, Academic Press, London, 1996, pp. 97-162. doi:10.1016/S0065-2296(08)60057-9
- J. Hidema, T. Kumagai, J. C. Sutherland and B. M. Sutherland, “Ultraviolet B-Sensitive Rice Cultivar Deficient in Cyclobuthyl Pyrimidine Dimer Repair,” Plant Physiology, Vol. 113, No. 1, 1997, pp. 39-44.
- J. Hidema, T. Kumagai and B. M. Sutherland, “UV Radiation-Sensitive Norin 1 Rice Contains Defective Cyclobutane Pyrimidine Dimer Photolyase,” Plant Cell, Vol. 12, No. 9, 2000, pp. 1569-1578.
- O. Danylchenko and B. Sorochinsky, “Use of RAPD Assay for the Detection of Mutation Changes in Plant DNA Induced by UV-B and γ-Rays,” BMC Plant Biology, Vol. 5, Suppl. 1, 2005, p. S9. doi:10.1186/1471-2229-5-S1-S9
- A. B. Britt, “DNA Damage and Repair in Plants,” Annual Review of Plant Physiology and Plant Moecular Biology, Vol. 47, 1996, pp. 75-100. doi:10.1146/annurev.arplant.47.1.75
- A. Sancar, “Structure and Function of DNA Photolyase,” Biochemistry, Vol. 33, No. 1, 1994, pp. 2-9. doi:10.1021/bi00167a001
- R. Beukers, A. P. M. Eker and P. H. M. Lohman, “50 Years Thymine Dimer,” DNA Repair, Vol. 7, No. 3, 2008, pp. 530-543. doi:10.1016/j.dnarep.2007.11.010
- F. E. Quaite, J. C. Sutherland and B. M. Sutherland, “Isolation of High-Molecular-Weight Plant DNA for DNA Damage Quantitation: Relative Effects of Solar 297 nm UVB and 365 nm Radiation,” Plant Molecular Biology, Vol. 24, No. 3, 1994, pp. 475-483. doi:10.1007/BF00024115
- F. E. Quaite, S. Takayanagi, J. Ruffini, J. C. Sutherland and B. M. Sutherland, “DNA Damage Levels Determine Cyclobutyl Pyrimidine Dimmer Repair Mechanism in Alfalfa Seedlings,” Plant Cell, Vol. 6, No. 11, 1994, pp. 1635- 1641.
- B. M. Sutherland, S. Takayanagi, J. H. Sullivan and J. C. Sutherland, “Plant Responses to Changing Environtmental Stress: Cyclobutyl Pyrimidine Dimmer Repair in Soybean Leaves,” Photochemistry and Photobiology, Vol. 64, No. 3, 1996, pp. 464-468. doi:10.1111/j.1751-1097.1996.tb03092.x
- S. T. Kim, K. Malhotra, C. A. Smith, J. S. Taylor and A. Sancar, “Characterization of (6-4) Photoproduct DNA Photolyase,” Journal of Biological Chemistry, Vol. 269, No. 11, 1994, pp. 8535-8540.
- A. Sancar, “Structure and Function of DNA Photolyase and Cryptochrome Blue-Light Photoreceptors,” Chemical Reviews, Vol. 103, No. 6, 2003, pp. 2203-2237. doi:10.1021/cr0204348
- C. Lin and T. Todo, “The Cryptochromes,” Genome Biology, Vol. 6, 2005, p. 220. doi:10.1186/gb-2005-6-5-220
- A. Sancar, “Structure and Function of Photolyase and in Vivo Enzymology: 50th Anniversary,” Journal of Biological Chemistry, Vol. 283, No. 47, 2008, pp. 32153-32157. doi:10.1074/jbc.R800052200
- M. Ahmad, J. A. Jarillo, L. J. Klimczak, L. G. Landry, T. Peng, R. L. Last and A. R. Cashmore, “An Enzyme Similar to Animal Type II Photolyases Mediates Photoreactivation in Arabidopsis,” Plant Cell, Vol. 9, 1997, pp. 199- 207.
- W. M. Waterworth, Q. Jiang, C. E. West, M. Nikaido and C. M. Bray, “Characterization of Arabidopsis Photolyase Enzymes and Analysis of Their Role in Protection from Ultraviolet-B Radiation,” Journal of Experimental Botany, Vol. 53, No. 371, 2002, pp. 1005-1015. doi:10.1093/jexbot/53.371.1005
- A. B. Britt, “Molecular Genetics of DNA Repair in Higher Plants,” Trends in Plant Science, Vol. 4, No. 1, 1999, pp. 20-25. doi:10.1016/S1360-1385(98)01355-7
- C. Z. Jiang, J. Yee, D. L. Mitchell and A. B. Britt, “Photorepair Mutants of Arabidopsis,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 94, No. 14, 1997, pp. 7441-7445. doi:10.1073/pnas.94.14.7441
- T. Hirouchi, S, Nakajima, T, Najrana, M. Tanaka, T. Matsunaga, J. Hidema, M. Teranishi, T. Fujino, T. Kumagai, and K. Yamamoto, “A Gene for a Class II DNA Photolyase from Oryza sativa: Cloning of the cDNA by Dilution-Amplification,” Molecular Genetic and Genomics, Vol. 269, No. 4, 2003, pp. 508-516. doi:10.1007/s00438-003-0856-9
- A. Yasui, A. P. M. Eker, S. Yasuhira, H. Yajima, T. Kobayashi, M. Takao and A. Oikawa, “A New Class of DNA Photolyases Present in Various Organisms Including Aplacental Mammals,” EMBO Journal, Vol. 13, No. 24, 1994, pp. 6143-6151.
- S. Kanai, R. Kikuno, H. Toh, H. Ryo and T. Todo, “Molecular Evolution of the Photolyase-Blue-Light Photoreceptor Family,” Journal of Molecular Evolution, Vol. 45, No. 5, 1997, pp. 535-548. doi:10.1007/PL00006258
- J. L. Johnson, S. Hamm-Alvarez, G. Payne, G. B. Sancar, K. V. Rajagopalan and A. Sancar, “Identification of the Second Chromophore of Escherichia coli and Yeast DNA Photolyases as 5,10-Methenyltetrahydrofolate,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 85, No. 7, 1988, pp. 2046-2050. doi:10.1073/pnas.85.7.2046
- T. Ueda, A. Kato, S. Kuramitsu, H. Terasawa and I. Shimada, “Identification and Characterization of a Second Chromophore of DNA Photolyase from Thermus thermophilus HB27,” Journal of Biological Chemistry, Vol. 280, No. 43, 2005, pp. 36237-36243. doi:10.1074/jbc.M507972200
- C. Lin, “Blue Light Receptors and Signal Transduction,” Plant Cell, Vol. 14, 2002, pp. S207-S225.

