American Journal of Plant Sciences
Vol.4 No.3(2013), Article ID:28987,6 pages DOI:10.4236/ajps.2013.43072
In Silico Analysis of a MRP Transporter Gene Reveals Its Possible Role in Anthocyanins or Flavonoids Transport in Oryze sativa*
![]()
1College of Life Science, Genetic Engineering Laboratory, South China Agricultural University, Guangzhou, China; 2College of Forestry, South China Agricultural University, Guangzhou, China; 3Vegetable Research Institute, Guangdong Academy of Agriculture Science, Guangzhou, China.
Email: #zhuql@scau.edu.cn
Received January 4th, 2013; revised February 8th, 2013; accepted February 16th, 2013
Keywords: In Silico Analysis; OsMRP15; a MRP Transporter; Anthocyanins or Flavonoids Transport; Oryze sativa
ABSTRACT
There are many studies on enzymatic pathways of anthocyanin biosynthesis, but little is known about the anthocyanins transport in Oryze sativa. In silico analysis, the OsMRP15 (LOC_Os06g06440), an orthologous gene of mazie anthocyanin transporter ZmMRP3, has been identified in rice. The OsMRP15 contained a 4425bp open reading frame (ORF) encoding a 1475 amino acid protein, belonging to a MRP subfamily of ABC transporters, and has a high sequence identity, very similar protein structure, and the same arrangement of domains to ZmMRP3, but the genomic structure of OsMRP15 was significant difference with ZmMRP3. The prediction promoter of OsMRP15 has many presumed anthocyanin regulatory sites. The phylogenetic analysis of MRPs in rice, mazie and Arabidopsis showed that OsMRP15 and ZmMRP3 belonged to the same subbranch. The expression pattern indicated that OsMRP15 was co-expression with two anthocyanin transcription factors. These analysis results implied that as an ortholog of ZmMRP3, the function of OsMRP15 was possibly as a membrane-bound transporter required for vacuolar uptake of anthocyanins in rice.
1. Introduction
The multidrug resistance-associated proteins (MRPs) belong to ATP-dependent, proton-gradient-independent transporters (ATP-Binding Cassette, ABC) superfamily [1], and the primary function of plant MRPs are involved in the vacuolar sequestration of potentially toxic metabolites, for example flavonoids and other secondary metabolites [2]. The full-size or typic MRP transporters consist of five core domains [3]: three hydrophobic transmembrane domains (TMDs) that each contains multiple transmembrane α-helices; and two soluble nucleotide binding domains (NBDs) that each is composed of the Walker A and B motives linked by an ABC signature motif. These domains are arranged with the topology: TMD0-TMD1- NBD1-TMD2-NBD2, in which the TMD0 is required for correct targeting and two (TMD-NBD)s compose a transmembrane transport channel.
Now, many MRPs have been identified in some different plants, but only several genes involved in flavonoids transport have been studied. There are 15 putative MRP genes in Arabidopsis genome. AtMRP1 and AtMRP2 transport anthocyanin-glutathione conjugates in vacuolar yeast [4,5]. Moreover, AtMRP2 is involved in chlorophyll degradation in vivo [6]. AtMRP4 and AtMRP5 are involved in guard cell regulation [7]. Maybe due to the function redundance in vacuolar uptake of anthocyanins, flavonoidless phenotypes have not been obatained by knockout mutants in some AtMRPs [2]. In mazie, ZmMRP3 is a membrane-bound anthocyanin transporter. The expression pattern of ZmMRP3 correlated with the anthocyanin accumulation pattern and is controlled by anthocyanin regulators [8]. ZmMRP4, the other related MRP, acts mainly as phytic acid transporter in the seed and its other possible function is partially redundant with ZmMRP3 in anthocyanin transport in the scutellum [9].
In rice, we identified 16 MRP genes and excluded the LOC_Os11g05700 that is a half transporter and consists of a single TMD and NBD, and originally called OsMRP16 in [3], using the latest RGAP V6.1 (Rice Genome Annotation Project) database, and only one gene was studied. OsMRP5 (LOC_Os03g04920, now-called OsMRP13 in this paper), an orthologus gene of ZmMRP4, is a phytic acid transporter in seeds [10].
Anthocyanin is an important biologically active substance in black, purple and red rice [11]. Although the regulators and key enzymes have been studied, little is known about anthocyanin transport in the final phase of anthocyanin biosynthesis in rice. In order to improve the content of anthocyanin in rice seeds, in this paper we identified a MRP gene, OsMRP15, and analyzed its possible role in anthocyanins or flavonoids transport.
2. Materials and Methods
2.1. Plant Materials
The Green rice varieties (ZH11, Nipponbare and T65) and red leaf variety ZY were planted in our lab.
2.2. RNA Isolation, cDNA Synthesis and RT-PCR Analysis
The total RNA was isolated by Trizol reagent kit (Invitrogen, USA) and two micrograms of total RNA were reverse-transcribed using M-MLV kit (Promega, USA), according to the manufacturer’s instructions. Using OsActin1 as a reference gene, two primers of OsMRP15, F15 (5’-agatgggtcgaattggagcatgggtc-3’) and R15 (5’-cgtaggtttgtcatac tccaccac-3’) were designed to RTPCR analysis, according to the procedure: predenaturation at 94˚C for 4 min, 35 cycles at 94˚C for 0.5 min, 58˚C for 0.5 min and 72˚C for 0.5 min, followed by the final extension at 72˚C for 5 min.
2.3. Bioinformatics Analysis
The latest RGAP V6.1 data was used to assay the rice MRP gene family. Gene prediction and structure analysis used a FGENESH web software. Molecular characterization and multiple sequence alignment of OsMRP15 was analyzed using Vector NTI 10.0 program (Invitrogen, USA). The promoter prediction and cis-acting regulatory elements analysis utilized the latest PromPredict program [12] and PLACE program [13], respectively. The conserved protein domain analysis used the SMART web tool, and protein subcellular localization prediction adopted WolF PSORT program. The peptide sequences of MRPs were aligned with Clustal W and subsequently a phylogenetic tree was constructed by the neighbor-joining (NJ) method with MEGA 5.0 by bootstrap with 1000 replicates [14]. Our Rice GeneChip Database was used to analyze gene expression patterns.
3. Results
3.1. Molecular Characteristics of OsMRP15
Using ZmMRP3 as a probe, the orthologus gene OsMRP15 was identified by BLAST search in RGAP V6.1 database. The OsMRP15 was located in Chr.6, and the TIGR number is LOC_Os06g06400. The genome length of OsMRP15 was 8445bp including 299bp 5’-UTR, 4425bp ORF and 389bp 3’-UTR. Although the gene CDS structure of OsMRP15 including 11 exons and 10 introns is very different with ZmMRP3 CDS structure that only contained 4 exons and 3 introns, the two genes have very similar phase of introns which implied that the introns loss maybe happened in ZmMRP3 (Figure 1). In addition, there were two introns in 5’-UTR of ZmMRP3, but no introns in prediction 5’-UTR of OsMRP15.
The OsMRP15 encoded a 1475 amino acid protein with a molecular weight of 164.51 kDa and a pI value of 7.69, belonged to a MRP subfamily of ABC transporters, and shared the high identity of 84.3% with ZmMRP3 protein. Multi-alignment of OsMRP15 with other plant MRPs showed high similarities. Like ZmMRP3, OsMRP15 was a full-size MRP transporter that has conserved TMD and NBD domains, and the domains arrangement were TMD0-TMD1-NBD1-TMD2-NBD2. The NBD domains are also characterized by Walker A motif (G-X(4)-G-K-[ST]), Walker B motif ([RK]-X(3)-G-X(3)- L-[hydrophobic]) and linked by an ABC signature motif ([LIVMFY]-S-[SG]-G-X(3)-[RKA]-[LIVMYA]-X-[LIV-MF]-[AG]) [7] (Figure 2).
The subcellular localization prediction of OsMRP15 showed its presence in the plasma membrane and was an integral membrane protein, which is similar with ZmMRP3 that located in the tonoplast [8]. And 48 conserved phosphorylation sites were predicted by NetPhos 2.0. These sites suggested that posttranslational modification may play an important role for OsMRP15 normal function. The protein structure analysis displayed that OsMRP15 and ZmMRP3 have similar second structure and almost exactly same of conserved structures except for lacking two transmembrane-spanning alpha-helices of TMD2 in ZmMRP3 (Figure 3).
3.2. The Analysis of Prediction Promoter Region of OsMRP15
In the 1.5 kb upstream of 5’-UTR of OsMRP15, a prediction core promoter region from −443bp to −503bp, many
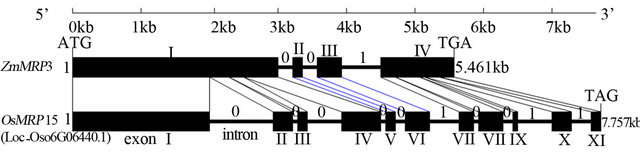
Figure 1. The genome structure of OsMRP15 compared with ZmMRP3. The number of 0 and 1 indicated the phase of introns: phase-0 introns (intron located between two codons), phase-1 introns (intron located between first and second nucleotide of a codon), respectively.

Figure 2. The deduced protein sequence and conserved structure of OsMRP15. The TMD0 was indicated by a dotted underline. The TMD1/TMD2 and NBD1/NBD2 were indicated by underlines and the boxes, respectively. The putative transmembrane-spanning alpha-helices were blue letter and shown in shadow. The Walker A and B motifs were indicated by red letter and bold letter in a grey background respectively, and the ABC signature motifs were shown in grey background.
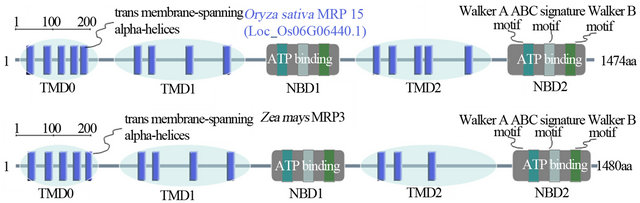
Figure 3. The protein structure comparison between OsMRP15 and ZmMRP3.
cis-active elements and many myb or myc cis-regulatory elements were found by using PromPredict and PLACE program, respectively (Figure 4). Moreover, a cis-sequence of OsMRP15 promoter similar to an anthocyanin regulatory element (ARE) [8], is present at −570 position (Figure 4). These promoter characteristics of OsMRP15 conformed to the ZmMRP3 promoter and implied that OsMRP15 expression was controlled by similar antho-cyanin regulatory genes.
3.3. The Phylogenetic Tree Analysis of OsMRP15
Using the latest RGAP V6.1 data, all 16 rice MRP genes were identified by BlASTP searches. The nomenclature for OsMRP1 to OsMRP7 and OsMRP9 to OsMRP15 were in accordance with [3]. The OsMRP8 was only one new locus not two in [3], and old OsMRP16 in [3] is a single TMD and NBD structure which was excluded in the rice MRP family. So, the new named OsMRP16 in this paper was the old OsMRP17.
In the phylogenetic tree (Figure 5), there were two clades: the clade I including two rice MRPs in which the possible function of OsMRP1 maybe was related with transport of anthocyanin-glutathione conjugates like AtMRP1 and AtMRP2 in yeast [4,5]; the clade II containing many subcludes in which OsMRP15 and ZmMRP3, and OsMRP13 and ZmMRP4 all belonged to the same subbranch with 100 bootstrap value, respectively. Therefore, it can be postulated that OsMRP15 was an anthocyanin transport similar with ZmMRP3, and OsMRP13 has possible redundant function in anthocyanin transport like ZmMRP4.
3.4. The Expression Pattern of OsMRP15
In our Rice GeneChip Database, OsMRP15 expression was higher in stigma than embryo, and very low in other tissues and organs. Furthermore, OsMRP15 was co-expression with two anthocyanin transcription factors, OsC1 and OsB1, in all tissues and organs, except for embryo (Figure 6). These expression patterns resembled ZmMRP3 [3] that expressed in all anthocyanin assembled and colored tissues and organs, and was controlled by the anthocyanin regulators B (myc type) and Pl (myb type).
In red leaf rice, the anthocyanin biosyntheses is controlled by OsC1 and OsB (OsB1 and OsB2) [15]. But in green leaf, the OsC1 is induced by UV-B and very low expression, and the OsB2 is non-function. Moreover, the anthocyanin key structure gene DFR is mutant and nonfunction in many japonica rice varieties, for example Nipponbare [15].
In order to detect the co-expression pattern of OsMRP15, the RT-PCR was analyzed in a red leaf variety ZY and 3 green leaf varieties ZH11, T65, and Nipponbare. The RT-PCR result indicated that OsMRP15
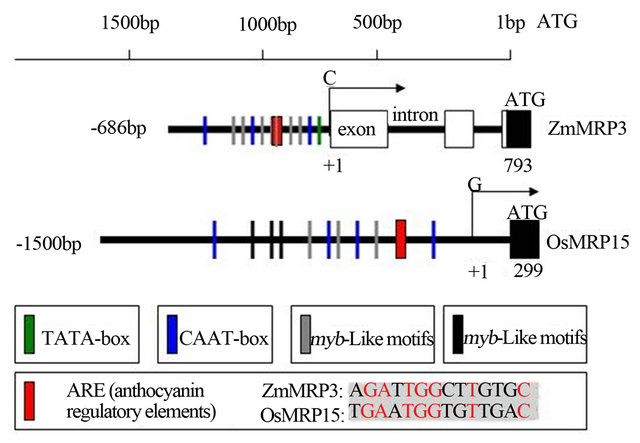
Figure 4. The promoter region comparison between OsMRP15 and ZmMRP3.
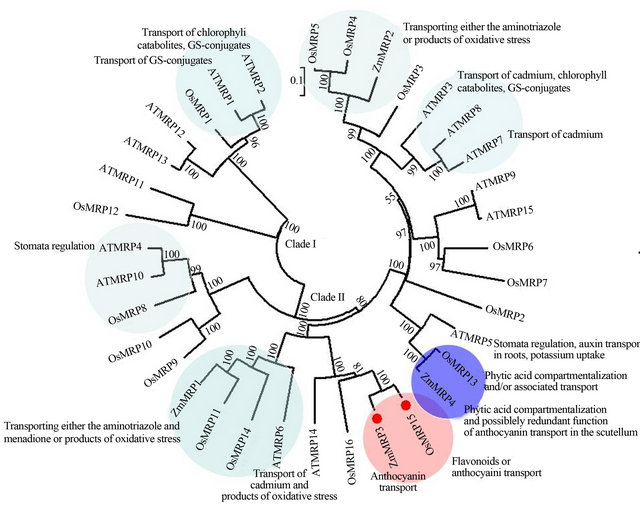
Figure 5. Phylogenetic tree of the MRP gene family in Arabidopsis, maize and rice. The OsMRP15 and ZmMRP3 were in the same subbranch indicated in pink background and others similar function genes were claded into different subclades in different colored background. The accession numbers of MRPs are: Arabidopsis and rice (OsMRP1 to OsMRP7 and OsMRP9 to OsMRP15) in accordance with [3]; OsMRP8: LOC_Os01g25386; OsMRP16: LOC_Os12g37580; ZmMRP1: AY186245; ZmMRP2: AY186247; ZmMRP3: AY609318.1; ZmMRP4: EF586878.
expression was co-expression with anthocyanin regulators in red leaf of ZY, but no expression in green leaf varieties (Figure 7). The integrative expression pattern suggested that OsMRP15 played a possibly important role in rice anthocyanin synthesis.
4. Discussion
As an ortholog of ZmMRP3 [8], a mazie anthocyanin transport, OsMRP15 was identified in silicon analysis using latest rice genome data. At the level of genome structure (Figure 1), it was very different that OsMRP15 has many exons in CDS and no introns in prediction 5’-UTR, but ZmMRP3 was only 4 exons in CDS and two introns in 5’-UTR. Compared with ZmMRP3, other orthologus structure were very similar with OsMRP15, which implied that introns have lost in ZmMRP3. On the other hand, at the level of protein, OsMRP15 has highest identity, the same protein structure, conserved domains and similar sublocation with ZmMRP3 (Figures 2 and 3). In prediction promoter of OsMRP15, many cis-active elements and cis-regulatory elements were very similar with ZmMRP3 promoter, specially the similar ARE [8].
The result of OsMRP15 and ZmMRP3 with high bootstrap value in the same subbranch of phylogenetic tree strongly suggested that the two genes have similar function. Interestingly, OsMRP16, the same subclade with OsMRP15, was no or very low expression in all tissue and organs (Figure 6), which implied that OsMRP16 maybe was a non-function gene in rice. The expression pattern of OsMRP15 co-expression with OsC1 and OsB1 was strongly supported by RT-PCR between red and green leaf rice varieties.
In green stigma, the function of OsMRP15 maybe was a flavonoids transport, in which transported some flavonoids, e.g. flavonols. Maybe because some key structure genes in anthocyanin biosynthesis were non-function or mutant, the pathway was incomplete though OsC1 and OsB1 were high expression. In addition, in embryo, OsMRP15 was expression but OsC1 and OsB1 were no expression. These results suggested that maybe OsMRP15 has other regulatory factors and played an unknown role in embryo, which was different with ZmMRP3.
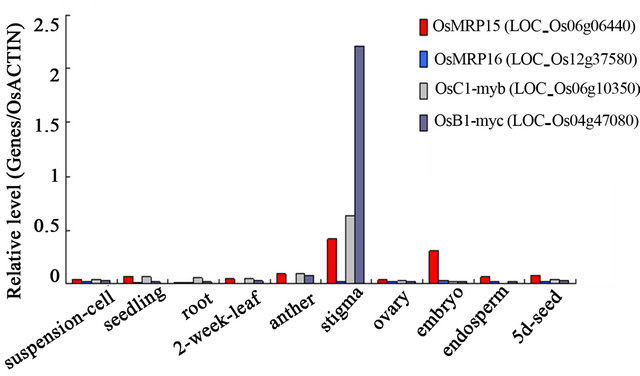
Figure 6. The expression patterns of OsMRP15 in different tissues and organs of rice. The relative level is the ratio of the signal intensity values between genes and OsACTIN1.

Figure 7. The RT-PCR analysis of OsMRP15 in green and red leaf rice cultivars.
Based on in silicon analysis and RT-PCR result, it can be deduced that OsMRP15 was a membrane-bound transporter that is required for vacuolar uptake of anthocyanins or flavonoids in rice, like ZmMRP3 in maize. Moreover, there was a possible GST gene presumptively forming anthocyanin-glutathione conjugates by OsMRP15 transporting into vacuolar in rice.
5. Acknowledgements
We thank Dr. Wu-Ming Xiao to provide the seeds of red leaf rice variety. We thank Ms. Elaine Zhou to amend the English language of the draft article.
REFERENCES
- M. Jasinski, E. Ducos, E. Martinoia and M. Boutry, “The ATP-Binding Cassette Transporters: Structure, Function, and Gene Family Comparison between Rice and Arabidopsis,” Plant Physiology, Vol. 131, No. 3, 2003, pp. 1169- 1177. doi:10.1104/pp.102.014720
- S. Kitamura, “Transport of Flavonoids: From Cytosolic Synthesis to Vacuolar Accumulation,” In: E Grotewold, Ed., The Science of Flavonoids, Springer, New York, 2006, pp. 123-146. doi:10.1007/978-0-387-28822-2_5
- M. Klein, B. Burla and E. Martinoia, “The Multidrug Resistance-Associated Protein (MRP/ABCC) Subfamily of ATP-Binding Cassette Transporters in Plants,” FEBS Letters, Vol. 580, No. 4, 2006, pp. 1112-1122. doi:10.1016/j.febslet.2005.11.056
- Y. P. Lu, Z. S. Li and P. A. Rea, “AtMRP1 Gene of Arabidopsis Encodes a Glutathione S-Conjugate Pump: Isolation and Functional Definition of a Plant ATP-Binding Cassette Transporter Gene,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 94, No. 15, 1997, pp. 8243-8248. doi:10.1073/pnas.94.15.8243
- Y. P. Lu, Z. S. Li, Y. M. Drozdowicz, S. Hörtensteiner, E. Martinoia and P. A. Rea, “AtMRP2, an Arabidopsis ATP Binding Cassette Transporter Able to Transport Glutathione S-Conjugates and Chlorophyll Catabolites: Functional Comparisons with AtMRP1,” Plant Cell, Vol. 10, No. 2, 1998, pp. 267-282.
- A. Frelet-Barrand, H. U. Kolukisaoglu, S. Plaza, M. Rüffer, L. Azevedo, S. Hörtensteiner, K. Marinova, B. Weder, B. Schulz and M. Klein, “Comparative Mutant Analysis of Arabidopsis ABCC-Type ABC Transporters: AtMRP2 Contributes to Detoxification, Vacuolar Organic Anion Transport and Chlorophyll Degradation,” Plant and Cell Physiology, Vol. 49, No. 4, 2008, pp. 557-569. doi:10.1093/pcp/pcn034
- B. Schulz and H. U. Kolukisaoglu, “Genomics of Plant ABC Transporters: The Alphabet of Photosynthetic Life Forms or just Holes in Membranes?” FEBS Letters, Vol. 580, No. 4, 2006, pp. 1010-1016. doi:10.1016/j.febslet.2006.01.002
- C. D. Goodman, P. Casati and V. Walbot, “A Multidrug Resistance-Associated Protein Involved in Anthocyanin Transport in Zea mays,” Plant Cell, Vol. 6, No. 7, 2004, pp. 1812-1826. doi:10.1105/tpc.022574
- F. C. Badone, E. Cassani, M. Landoni, E. Doria, D. Panzeri, C. Lago, F. Mesiti, E. Nielsen and R. Pilu, “The Low Phytic Acid 1-241 (Lpa1-241) Maize Mutation Alters the Accumulation of Anthocyanin Pigment in the Kernel,” Planta, Vol. 231, No. 5, 2010, pp. 1189-1199. doi:10.1007/s00425-010-1123-z
- X. H. Xu, H. J. Zhao, Q. L. Liu, T. Frank, K. H. Engel, G. An and Q. Y. Shu, “Mutations of the Multi-Drug Resistance-Associated Protein ABC Transporter Gene 5 Result in Reduction of Phytic Acid in Rice Seeds,” Theoretical and Applied Genetics, Vol. 119, No. 1, 2009, pp. 75-83. doi:10.1007/s00122-009-1018-1
- H. Ichikawa, T. Ichiyanagi, B. Xu, Y. Yoshii, M. Nakajima and T. Konishi, “Antioxidant Activity of Anthocyanin Extract from Purple Black Rice,” Journal of Medicinal Food, Vol. 4, No. 4, 2001, pp. 211-218. doi:10.1089/10966200152744481
- C. Morey, S. Mookherjee, G. Rajasekaran and M. Bansal, “DNA Free Energy-Based Promoter Prediction and Comparative Analysis of Arabidopsis and Rice Genomes,” Plant Physiology, Vol. 156, No. 3, 2011, pp. 1300-1315. doi:10.1104/pp.110.167809
- K. Higo, Y. Ugawa, M. Iwamoto and T. Korenaga, “Plant CisActing Regulatory DNA Elements (PLACE) Database: 1999,” Nucleic Acids Research, Vol. 27, No. 1, 1999, pp. 297-300. doi:10.1093/nar/27.1.297
- K. Tamura, D. Peterson, N. Peterson, G. Stecher, M. Nei and S. Kumar, “MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods,” Molecular Biology and Evolution, Vol. 28, No. 10, 2011, pp. 2731- 2739. doi:10.1093/molbev/msr121
- C. H. Shih, H. Chu, L. K. Tang, W. Sakamoto, M. Maekawa, I. K. Chu, M. Wang and C. Lo, “Functional Characterization of Key Structural Genes in Rice Flavonoid Biosynthesis,” Planta, Vol. 228, No. 6, 2008, pp. 1043- 1054. doi:10.1007/s00425-008-0806-1
NOTES
*Sponsor: Project 31000698 supported by National Natural Science Foundation of China.
#Corresponding author.

