Advances in Bioscience and Biotechnology
Vol.4 No.3(2013), Article ID:28780,5 pages DOI:10.4236/abb.2013.43051
Senescence process and oxidative stresses induce changes in plant genomic DNA quality
![]()
1Research & Production Complex, Pasteur Institute of Iran, Tehran, Iran
2Department of Agronomy and Plant Breeding, College of Aburaihan, University of Tehran, Pakdasht, Iran
Email: *mghorbani@irimc.org
Received 2 January 2013; revised 5 February 2013; accepted 25 February 2013
Keywords: RAPD; Senescence; Oxidative Stress; Genetically Erosion
ABSTRACT
Senescence or programmed cell death is a process that interacts with many biochemical and physiological changes in living organism and is generally induced by aging. Many environmental stresses that accelerate the production of activated oxygen can also induce senescence artificially. One of the important aspects of senescence is possibly degradation of macromolecules such as DNA. It is believed that the random amplification of polymorphic DNA (RAPD) technique is a good method to compare the DNA quality of juvenile and senescence samples in which oxidative stress is induced. In this study, juvenile, senescence and plant paraquat treated leaves from tomato, tobacco and rose, as well as juvenile and senescence human tissues were processed for DNA extraction followed by RAPD technique. We discovered that plant and human genomes are influenced by senescence and environmental stresses underwent genome diversity. Using some molecular markers proved that senescence and oxidative treated samples show different DNA pattern compare to the juvenile-untreated samples. We also concluded that RAPD technique can be used as a useful tool in genomics study to provide researchers reliable information of DNA quality and can effectively help to resolve the environment condition.
1. INTRODUCTION
Random amplification of polymorphic DNA (RAPD) is a technique using single, short (usually 10-mer) synthetic oligonucleotide perimers for PCR, the primer, whose sequence has been chosen randomly [1]. In fact RAPD is PCR based techniques developed by Williams et al. [2]
and has been initially used to detect polymorphism in genetic mapping, spouse taxonomy and phylogenetic studies and later in genotoxicity and carcinogenesis and other damages that is related to DNA quality [3]. In comparison to other fingerprinting methods, after proper optimization, the RAPD is a reliable, fast, sensitive and reproducible assay that allows it to detect a wide range of DNA damages [4,5]. RAPD technique has been shown to have the ability to detect the minimal quantity of DNA, assess the multiple regions throughout the genome and allow rapid analysis of large quantity of DNA. RAPD can provide specific detail of DNA damage or the genome sequence as well as detecting radioactive or enzymatic degradation of PCR product prior the analysis [6, 7].
Damage or change in DNA might be induced by many factors. Many researchers have reported that in the senescence or programmed cell death, defined as deteriorative processes, there are natural causes of death and degradation of macromolecules such as DNA [8,9]. Reports have also shown that many oxidative stresses such as paraquat treatment can artificially induce senescence. Paraquat is the member of the bipyridylium herbicides (1,1’dimethyl-4,4’bipyridylium) that act on the electron chain reaction on the chloroplast or mitochondria to produce reactive oxidative singles (ROS) [10,11].
In fact, the common point between oxidative stresses such as paraquat and senescence is increasing the production of ROS in different cell organelles. It has been suggested that excess of ROS in this process may induce damages such as DNA degradation [8,12]. Imlay and Linn discovered that both the sugar and the base moieties in DNA structure are susceptible to oxidation, causing base degradation, single strand breakage, and crosslinking to protein. Reactive oxygen have many direct and indirect effects on rubisco [13].
Investigations on herbicides mechanisms have shown that reactive oxygen radicals can directly induce rubisco fragmentation to 37 and 16 KD polypeptides. Ishida et al. reported that reactive oxygen species may triggers the site specific degradation of the large subunit (LSU) of rubisco in lysates of chloroplast exposed to light [14]. In 1996 Desimone et al. demonstrated that oxidative stresses could induce partial degradation of rubisco in isolated chloroplast of barley [15]. Liu et al. also found that the large subunit of rubisco is cleaved by reactive oxygen spicies at the metal binding site, close to the active site of the molecule [7].
CND41 is a protease from Aspartate proteases family which was discovered first by Nakano in 1997 [16]. WHO also identified the cDNA sequence of this protein from its amino acid sequence and reported that CND41, a DNA binding aspartic protease in chloroplast, has an important role in rubisco degradation in senescence and by decreasing the expression of CND41 in transgenic tobacco, senescence will be delayed. There are a few proteins in this family and all posses a homeobox containing a short (180 bp) conserved DNA sequence that encodes a DNA-binding motif known for its presence in genes that are involved in orchestrating the development of a wide range of organism. The presence of CND41 genome in different plants such as tobacco, tomato, roses, grape as well as human and mouse genome is well documented in literatures. It appears that the homeobox and partial sequence of CND41 has a role in aging and it is suggested that it is a conserved region in genes causing aging. Dalling et al. were the first group to suggest that oxidative modification of rubisco may target CND41 protein for subsequence degradation [17]. More investigation about action of this protease has shown that CND41 can only have proteolytic activity on deactivated rubisco [18].
Landry and Pell noted that ozone may induce oxidative modification of Rubisco leading to subsequent proteolysis [19]. Eckardt and Pell also reported that oxygen free radicals resulted from O3 exposing, caused enhanced degradation of rubisco in mature leaves of Solanum tuberosum L. [20].
Zhou et al. in 2006 indicated that H2O2 accumulation in chloroplast was negatively correlated with the initial rubisco activity and photosynthetic rate [21]. The reactive oxygens may cause the cleaves of the large subunit or modify rubisco to become more susceptible to proteolysis [22].
Reactive oxygen may also indirectly affect other macromolecules resembling DNA and proteins and induce more activation of rubisco specific proteases in the translational or post translational stage of expression that lead to more rubisco degradation. In 2004 Kato et al. found that the reduction of DNA copy numbers during senescence could initiate the activation of CND41 as a protease [23]. The objective of this project was to quantifying the special effects of oxidative burst on the activetion and content of rubisco in result of senescence and oxidative paraquat stress. For this reason we examined the rubisco content.
In this study, we investigated the DNA damages due to senescence and aging in human and plant samples as well as the effects of paraquat oxidative treatment on plant DNA quality by using RAPD technique.
2. MATERIAL AND METHOD
2.1. Plant Material and Sample Preparation
Plants including tomato, tobacco and rose were grown in greenhouse under controlled condition. After well establishment of plants, distinct leaves at middle part of each plant were sprayed thoroughly with different paraquat (methyl viologen, 1,1’-dimethyl-4,4’-bipyridinium dichloride; Sigma, St Louis, MO, USA) doses (0, 300 µM, 600 µM) diluted in 4% Tween 80 in distilled water and exposed to light for 3 hours. The control leaves was only sprayed with 4% Tween in distilled water and exposed to light for 3 hours as well. To determine DNA damages along with senescence in plants, juvenile (perfectly green) and senescence (yellow) leaves of each plant were used.
2.2. DNA Extraction
Total DNA of plant samples were extracted according to the Cetyl Trimethyl Ammonium Bromide (CTAB) activated charcoal protocol of Krizman et al. (2006) with some modifications [24]. To inhibit DNA degradation during extraction, fresh leaf samples (0.5 g) were cut by blade to small fragments (1 × 1 mm). After adding 0.6 g CTAB, 0.03 g PVP and 0.015 g activated charcoal, leaves were softly homogenized in 3 ml extraction buffer contain 100 mM Tris-HCl (pH 8), 2.0 M NaCl, 20 mM EDTA (pH 8). The homogenate was then transferd into a microcentrifuge tube and incubated at 55˚C for 30 min with frequent agitation, avoiding the suspension to settle. It was then cooled down to room temperature and centrifuged at 16,000 g for 10 min at room temperature. After the transfer of supernatant to a new tube, 1 volume of chloroform-isoamylalcohol (4% (v/v)) was added to the supernatant and vortexed thoroughly. Then, Centrifuged at 16,000 g for 10 min at room temperature and the aqueous (upper) phase was transferred to a new tube. If cloudiness in the solution persists, the chloroform-isoamylalcohol extraction should be repeated again. The supernatant was then transferred to a new tube and 0.45 volume of isopropanol was added and mixed by inversion. The mixture was incubated at 25˚C for 1 hour and centrifuged at 700 g for 10 min at room temperature. The supernatant was discarded and the remaining pellet was washed by adding 1 mL of wash buffer followed by vortexing. After centrifugation at 900 g for 10 min at room temperature and discarding the supernatant the pellet was air dried and re-suspended in 50 µL of distilled water.
2.3. Human Material and DNA Extraction
To assay DNA changes during senescence in human and avoid the side effects of other DNA changes that are unique in every person, black and white hairs of one person were selected. Human DNA extraction was done according to the Sambrook and Russell protocol 6 of molecular cloning (2001) [25].
2.4. Amplification of DNA
RAPD-PCR primers were designed as C08, E07 and P06 (Table 1). All reactions were performed with 35 cycles and annealing temperature of 35˚C. The extension and denaturing temperatures were set at 72˚C for 1 min and 94˚C for 30 seconds as quoted in most standard protocols. RAPD-PCR products were then separated in 2% agarose gel due to small size of the products and stained with ethidium bromide. To amplify CND41 gene (NCBI, D26015), forwards primers, CNF1 and CNF2, and reverse primers, CNR1 and CNR2, applied (Table 1).
2.5. CND41 Gene Expression Using HPLC
High-performance liquid chromatography (HPLC) was performed using (H2O: Acetonitrile with formic acid) on λ 215 nm column in C18 at 1 ml/min and 25˚C at before and after paraquat treatment and in juvenile and senescence leaves.
3. RESULTS
As shown in Figure 1, RAPD profile of Juvenile, senescence and paraquat treated samples of each plant (tomato, tobacco and rose) is completely different. Two unknown bands with ~300 bp and ~400 bp length appeared in senescence and paraquat treated of tomato, respectively, whereas there are no such bands in juvenile tomato plant. Approximately the same pattern is present in cases of tobacco and rose plants, but with the presence of different band sizes (Figure 1). Different RAPD profile using each primer might be the consequence of genome breakage.
Using RAPD markers, we demonstrated that the pattern of DNA quality of human samples was similar to plant samples indicating a variation in band intensity, loss of some bands and appearance of new bands in comparison of RAPD profiles of white and black hair of one subject (Figure 2).
We have also observed the presence of these sequences in the genome of the plant species including buxus, grapes and animal genomes such as mice (data not shown). It is interesting that the amplified sequences using specific primers showed that they belong to a catalytic enzyme that is present in the human genome (Figure 3).
HPLC results showed that there are two maximum extents in all treatments chromatogram. This is to be expected that the first maximum of chromatogram is related to Rubisco because of its high amount as compared to other proteins. The second maximum that is characterized after 10 minutes is coincidence with predicted CND41 enzyme (Figure 4).
4. DISCUSSION
In this experiment, assessing and comparison of the DNA quality of juvenile, senescence and paraquat treated samples of plant showed that although they were from plants with same genetic background, their RAPD profiles were somewhat different, mainly because of the differences in their age and environmental condition. It also appears that oxidative stresses may induce some changes in DNA stability.
Investigation of the DNA quality of human samples (white and black hair) as well showed the same results.
Difference in RAPD profiles including variation in band intensity, disappearance of bands, and appearance of new PCR products could be due to genetic changes as a result of stress due to environmental changes.
Modification of band intensity and loss of bands could also be due to alteration of the site of primer bindings or site of interaction of DNA polymerase with DNA [7]. Loss of band or their intensity could also be referred to the damage of DNA template.
Table 1. PCR primers used in this study for random amplification of genome and specific amplification of CND41.
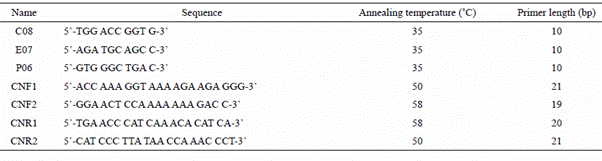
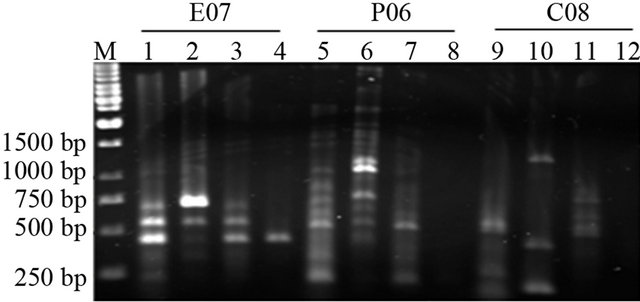
Figure 1. RAPD profile of Juvenile (1, 5 and 9 lanes), senescence (4, 8 and 12), 300 µM of paraquat (2, 6 and 10 lanes), 600 µM of paraquat (3, 7 and 11 lanes) treated samples of tomato using arbitrary primers. First four lanes are product of Primer E07, second four P06 and third four lanes are product of C08 primer.
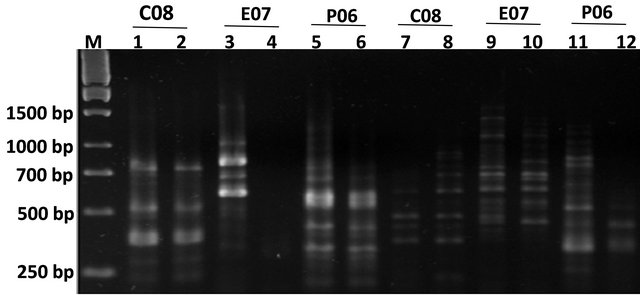
Figure 2. Agarose gel electrophoresis for PCR products of juvenile, senescence and paraquat treated plant samples as well as human samples. Lanes 1 and 2 represent juvenile and paraquat treated tobacco PCR samples respectively with CO8 primers. Lanes 3 and 4 represent juvenile and paraquat treated tobacco PCR samples respectively with E07 primers. Lanes 5 and 6 represent juvenile and paraquat treated tobacco PCR samples respectively with P06 primers. The next 6 lanes represent human black and white hairs in the same order as previous 6 lanes including the primer orders.
Figure 3. Agarose gel electrophoresis for PCR products of tobacco and human samples. (a) Lanes 1and 2 represent tobacco and human PCR samples respectively with CNF2 and CNR1 primers; (b) Lanes 1 and 2 represent tobacco and human PCR samples respectively with CNR2 and CNF1 primers.

Figure 4. Agarose gel electrophoresis for PCR products of juvenile, senescence and paraquat treated plant samples as well as human samples. Lanes 1 and 2 represent juvenile and paraquat treated tobacco PCR samples respectively with CO8 primers. Lanes 3 and 4 represent juvenile and paraquat treated tobacco PCR samples respectively with E07 primers. Lanes 5 and 6 represent juvenile and paraquat treated tobacco PCR samples respectively with P06 primers.
Appearance of new bands with higher molecular weight or increasing the density of heavier bands may refer to more access of DNA polymerase to the DNA template or alteration of the location of primer binding site and placing more DNA template between forward and reverse primer. Appearance of new PCR product also may occur because some oligonucleotide priming site could become accessible to oligonucleotide primers after structural changes or because some changes in DNA sequence have occurred due to mutations (resulting in new annealing events) or large deletions (bringing two pre-existing annealing site closer), or homologous recombination (juxtaposing two sequence that match the sequence of primer) [7].
Although it has been shown that changing the color of hair during senescence is a kind of differentiation process, changes between RAPD profile were approximately resembled the differences of the RAPD profile of juvenile, senescence and paraquat treated samples of plant, therefore, variation in band intensity, loss of some bands and appearance of new bands in comparison were seen.
Since the amplified genes belong to the catalytic enzymes, it appears that the presence of this gene in both plant and animal genomes could belong to homeobox sequences with proteolytic activity and responsible for aging in both plants and animals. The existence of the mRNA sequence of CDN41 gene in many plant and animal genomes is the evidence of conservation of this gene in eukaryote cells. Nakano et al. were succeeded to identify and synthesis the cDNA of CDN41 gene from its amino acid sequence and demonstrated that the deletion of this gene could delay aging in transgenic tobacco plant and reported that CDN41 protein was involved in both aging and catalysis of Rubisco [17]. We conclude that the presence of CDN41 gene in human could be responsible for aging and inhibition of the activity of this enzyme or decreasing the expression of this protein could delay aging in human.
![]()
![]()
REFERENCES
- Arora, S.K., Wolfgang, M.C., Lory, S. and Ramphal, R.J. (2004) Sequence polymorphism in the glycosylation island and flagellins of Pseudomonas aeruginosa. Bacteriology, 186, 2115-2122.
- Williams, J.G., Kubelik, A.R., Livak, K.J., Rafalski, J.A. and Tingey, S.V. (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research, 18, 6531-6535.
- Torezan J.M.D., Souza R.F.D., Ruas P.M., Ruas C.D., Camargo E.H and Vanzela A.L.L. (2005) Genetic variability of pre and post-fragmentation cohorts of Aspidosperma polyneuron Muell. Arg. (Apocynaceae). Brazilian Archives of Biology and Technology, 48, 171-180.
- Atienzar, F.A. and Jha, A.N. (2006) The random amplified polymorphic DNA (RAPD) assay and related techniques applied to genotoxicity and carcinogenesis studies: A critical review. Mutation Research/Reviews in Mutation Research, 613, 76-102. doi:10.1016/j.mrrev.2006.06.001
- Corich, V., Mattiazzi, A., Soldati, E., Carraro, A. and Giacomini, A. (2005) Sau-PCR, a Novel Amplification Technique for Genetic Fingerprinting of Microorganisms. Applied and Environmental Microbiology, 71, 6401-6406. doi:10.1128/AEM.71.10.6401-6406.2005
- Siwoski, A., Ishkanian, A., Garnis, C., Zhang, L., Rosin, M. and Lam, W.L. (2002) An efficient method for the assessment of DNA quality of archival microdissected specimens. Modern Pathology, 15, 889-892. doi:10.1097/01.MP.0000024288.63070.4F
- Liu, C., Bai, B., Skogerbø, G., Cai, L., Deng, W., Zhang, Y., Bu, D., Zhao, Y. and Chen, R. (2005) NONCODE: An integrated knowledge database of non-coding RNAs. Nucleic Acids Research, 33, D112-D115.
- Ranjan, R., Marczewski, A., Chojnacki, T., Hertel, J. and Swiezewska, E. (2001) Search for polyprenols in leaves of evergreen and deciduous Ericaceae plants. Acta Biochimica Polonica, 48, 579-584.
- Khanna-Chopra, R. (2012) Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation. Protoplasma, 249, 469-481. doi:10.1007/s00709-011-0308-z
- Donahue, J.L., Okpodu, C.M., Cramer, C.L., Grabau, E.A. and Alscher, R.G. (1997) Responses of antioxidants to paraquat in pea leaves (relationships to resistance). Plant Physiology, 113, 249-257.
- Shaaltiel, Y. and Gressel, J. (1987) Evidence that paraquat transiently inhibits leaf chloroplast reactions in resistant plants. Plant Physiology, 85, 869-871. doi:10.1104/pp.85.4.869
- Kryston, T.B., Georgiev, A.B., Pissis, P. and Georgakilas, A.G. (2011) Role of oxidative stress and DNA damage in human carcinogenesis. Mutation Research, 711, 193-201.
- Imlay, J.A. and Linn, S. (1986) DNA damage and oxygen radical toxicity. Science, 240, 1302-1309. doi:10.1126/science.3287616
- Ishida, H., Makino, A. and Mae, T. (1999) Fragmentation of the large subunit of ribulose-1,5-bisphosphate carboxylase by reactive oxygen species occurs near Gly-329. Journal of Biological Chemistry, 274, 5222-5226.
- Desimone, M., Henke, A. and Wagner, E. (1996) Oxidative stress induces partial degradation of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase in isolated chloroplasts of barley. Plant Physiology, 111, 789-796.
- Nakano, T., Murakami, S., Shoji, T., Yoshida, S., Yamada, Y. and Sato, F. (1997) A novel protein with DNA binding activity from tobacco chloroplast nucleoids. Plant Cell, 9, 1673-1682.
- Dalling, M.J., Hucklesby, D.P. and Hageman, R.H. (1987) A comparison of nitrite reductase enzymes from green leaves, scutella, and roots of corn (Zea mays L.). Plant Physiology, 51, 481-484. doi:10.1104/pp.51.3.481
- Kato, Y., Yamamoto, Y., Murakami, S. and Sato, F. (2005) Post-translational regulation of CND41 protease activity in senescent tobacco leaves. Planta, 222, 643-651. doi:10.1007/s00425-005-0011-4
- Landry, L.G. and Pell, E.J. (1993) Modification of Rubisco and Altered Proteolytic Activity in O3-Stressed Hybrid Poplar (Populus maximowizii x trichocarpa). Plant Physiology, 101, 1355-1362.
- Eckardt, N.A. and Pell, E.J. (1996) Effects of ethylenediurea (EDU) on ozone-induced acceleration of foliar senescence in potato (Solanum tuberosum L.). Environmental Pollution, 92, 299-306. doi:10.1016/0269-7491(95)00111-5
- Zhou, Y.H., Yu, J.Q., Mao, W.H., Huang, L.F., Song, X.S. and Nogués, S. (2006) Genotypic variation of rubisco expression, photosynthetic electron flow and antioxidant metabolism in the chloroplasts of chill-exposed cucumber plants. Plant and Cell Physiology, 47, 192-199. doi:10.1093/pcp/pci234
- Feller, U., Anders, I. and Mae, T. (2008) Rubiscolytics: Fate of Rubisco after its enzymatic function in a cell is terminated. Journal of Experimental Botany, 59, 1615- 1624. doi:10.1093/jxb/erm242
- Kato, Y., Murakami, S., Yamamoto, Y., Chatani, H., Kondo, Y., Nakano, T., Yokota, A. and Sato, F. (2004) The DNA-binding protease, CND41, and the degradation of ribulose-1,5-bisphosphate carboxylase/oxygenase in senescent leaves of tobacco. Planta, 220, 97-104. doi:10.1007/s00425-004-1328-0
- Krizman, M., Jakse, J., Baricevic, D., Javornik, B. and Prosek, M. (2006) Robust CTAB-activated charcoal protocol for plant DNA extraction. Acta Agriculture Slovenica, 87, 427-433.
- Sambrook, J. and Russell, D.W. (2001) Molecular cloning: A laboratory manual. Vol. 1, Cold Spring Harbor Laboratory Press, New York.
NOTES
*Corresponding author.

