Modulation of Specific Apoptotic DNA Fragmentation after Short Term Exposure to Natural UVR in Fish Larvae ()
1. Introduction
Ultraviolet radiation (UVR) is known to cause damages in aquatic organisms [1] , eventually leading to increased mortality [2] . An exposure to UVR may result in sunburn, growth impairment and injuries to brain tissue and retina in fishes [3] . Most studies focused on UV-B (280 - 315 nm), showing that these wavelengths can cause cyclobutane pyrimidine dimers (CPD) formation in deoxyribonucleic acid (DNA) [4] -[6] . These damages may cause apoptotic cell death, also known as programmed cell death, as reported in studies on Atlantic cod larvae [7] . Indeed, like other stressors (i.e., toxins), UVR can initiate the apoptotic cascade through different pathways, all resulting in DNA fragmentation and cell removal [8] [9] . The two main apoptotic pathways, namely the intrinsic and extrinsic one, are respectively activated through an intracellular injury or by the activation of death receptors on the cellular membrane [10] . On the other side, apoptosis is also the most important mechanism in preand post-natal development, allowing suppression of damaged, unwanted or badly located cells, as reported by extensive works on zebrafish (Danio rerio) [11] -[13] . Therefore, whereas a basal rate of apoptosis is normally observed in developing organisms such as fish larvae going through morphogenesis, external stressors such as UVR may affect developmental apoptosis by modulating specific rates of cellular death [7] .
The role of UV-A in inducing apoptosis is not as clear as for the UV-B, while UV-A wavelengths can cause photodammages to DNA, they can also induce DNA photoreparation [14] [15] . Similarly to UV-B, UV-A may cause CPD formation in DNA [14] [16] , as well as reactive oxygen species (ROS) generation [17] . A relatively high ROS/antioxidants ratio in the cell can lead to oxidative stress, which can damage proteins, lipids and DNA [18] , and eventually drive the cell towards apoptosis and death. On the other hand, UV-A and visible light may counteract these effects by activating the CPD photolyase enzyme [15] [19] , which directly repairs CPD damages.
Most studies on the responses of aquatic organisms to UVR-induced apoptosis were performed under artificial light and/or in laboratory controlled environments. Yet, UVR lamps are generally biased towards UV-B and do not adequately reproduce realistic UV-A/UV-B ratios and consequently tend to overestimating photodamages. UVA-induced photorepair is indeed reduced in laboratory conditions, and experimental results tend to exaggerating the effects of UVR [20] [21] . On the other hand, in studies reporting data under natural UVR, the apoptosis parameters were not measured [20] [22] [23] . Thus, the goal of our study was to measure the effects of spectral quality (visible light, visible light + UV-A or visible light + UV-A + UV-B) and intensity (high, medium, or low) of natural UVR on several apoptosis markers in fish larvae. More specifically, we measured specific apoptotic DNA fragmentation as well as the expression of proand anti-apoptotic proteins, p53, Bax and Bcl-2, and the modulation of PARP-1, after two days of exposure to nine different natural solar light treatments (complete 3 × 3 experimental factorial plan). Yellow perch larvae (Perca flavescens) were chosen as model species because previous experimental studies showed that natural levels of UVR could induce mortality in both embryos [24] and larvae [23] of this species.
Since our study was performed under natural solar light, our results were expected to allow for a more accurate and realistic response of the impact of UVR in the balance underlying developmental apoptosis and stressorinduced apoptosis in aquatic organisms.
2. Material and Methods
2.1. Material
All chemicals were purchased from Sigma (St. Louis, MO) unless specified otherwise.
2.2. Egg Sampling
The Yellow perch eggs were sampled on 15 April 2010 at Milieu Island, on the north shore of Sorel-Berthier archipelago, St. Lawrence River (Quebec, Canada) (46˚04'52.06"N, 73˚10'00.98"O). Eggs were incubated in aquariums of the Aquatic Communities Research Laboratory of Université du Québec à Trois-Rivières at temperatures between 18˚C and 20˚C. Egg strands were gently shacked to help hatching and start the experiments with larvae at the same development phase. After the resorption of the yolk sack, larvae were fed with Tetramin® flakes for young fishes.
2.3. Larvae Exposure to Natural Solar Light
Groups of 45 subjects were randomly formed with freshly hatched larvae. The larvae were placed in incubators which consisted of plastic transparent containers (6 cm depth, 11 cm diameter) with a ~200 µm mesh size net on the bottom. A total of 20 incubators were equally distributed in two Styrofoam® coolers. Incubators were randomized in each cooler, the latter representing a “block”.
Three different selective filters were used to modulate spectral quality of light: Whirlpak® (Nasco, Atkinson, WI) for full solar spectrum (hereafter called “visible light + UV-A + UV-B”) (Figure 2 [23] ), Mylar-d® (SABIC polymershapes, Montréal, QC, Canada), opaque to UV-B (hereafter called “visible light + UV-A”) and J-Roll® (SABIC polymershapes, Montréal, QC, Canada), opaque to UV-A and UV-B (hereafter called “visible light”). The Whirlpak® filter (UV-B + UV-A + visible light) was highly transparent to UVR: on average, 83.57% ± 1.82% (mean ± SD) of UV-B (wavelengths between 280 and 320 nm) and 88.01% ± 1.06% of UV-A (wavelengths between 321 and 400 nm) passed thought it (Figure 1). Mylar-d® (UV-A + visible light) had a mean transmittance of 82.61% ± 3.93% for UV-A and of 7.64% ± 16.93% for UV-B; this filter nearly completely cuts off UV-B <310 nm. Jroll® (visible light) had a mean transmittance of 0.00% ± 0.30 for UV-B and 6.90% ± 12.57% for UV-A. The three selective filters were essentially transparent to visible light (wavelengths between 400 and 700 nm): the transmittances of the Whirlpak®, Mylar-d® and Jroll® were of 90.24% ± 0.44%, 88.04% ± 0.61% and 87.03% ± 4.21%, respectively (Figure 2 in [23] ). To modulate the intensity of light, three levels of neutral density filters (window screens: none, one or two) were installed on the selective filters [25] . One sheet of window screens blocked 38.40% ± 0.47% of total irradiance at each wavelength while two sheets blocked 66.23% ± 0.56% [23] . The three levels of wavelength filters were crossed with three levels of neutral filters to get a complete factorial plan (3 × 3). The control condition was obtained by covering incubators with an aluminium foil (hereafter called “dark”), which is useful to rule out whether the visible light treatment plays a role on apoptosis.
Larvae were exposed to complete solar spectrum for two days (20 and 21 April 2010, which were two sunny days) on the Université du Québec à Trois-Rivières campus (Québec, Canada; 46˚35'N, 72˚58'O).
Samples of living larvae were taken at the end of the second day of the experiment. The larvae were euthanized with carbonated water, and all samples were frozen in 30% (w/v) sucrose solution at −80˚C to prevent the breaking of cell membranes.
During the experimental period, the surface irradiance was measured with a PUV-2545/2546 radiometer (Biospherical Instruments Inc., San Diego, CA) at a frequency of 5 Hz, wavelengths of 313 nm and 320 nm for UV-B, 340 nm for UV-A and 400 - 700 nm for visible light. Percent transmission of each selective filter was calculated using the radiometer at the four wavelengths mentioned above from the ratio of the irradiance under

Figure 1. Effect of UVR quality and intensity on yellow perch larvae survival. Dark grey: Whirlpak® (visible + UV-A + UV-B), light grey: Mylar-d® (visible + UV-A), and white: J-Roll® (visible). Box plots show median values with the 25th and 75th percentiles. The bars represent the range of observed values that fall within 1.5 times the interquartile range (25% - 50%). None of the treatments influences larvae survival after two days of exposure.

Figure 2. Effect of UVR quality and intensity on yellow perch larvae apoptosis. Dark grey: Whirlpak® (visible + UV-A + UV-B), light grey: Mylar-d® (visible + UV-A), and white: J-Roll® (visible). Specific apoptotic DNA fragmentation was measured in function of received light treatment. Specific apoptotic DNA fragmentation changes in function of light quality: treatments with UV-A + visible or UV-B + UV-A + visible reduce apoptotic DNA fragmentation.
the filter (measured with the radiometer covered with the different plastic films) and the total irradiance (measured with the uncovered radiometer) (see Figure 2 in [23] ).
2.4. Detection of Specific Apoptotic DNA Fragmentation
Specific detection of apoptotic DNA fragmentation was studied using the ssDNA Apoptosis ELISA kit (Chemicon International, Billerica, MA), as already reported [26] . This procedure is based on the selective denaturation of DNA by formamide in apoptotic cells but not in necrotic cells [27] . Importantly, this technique specifically enables the identification of apoptotic DNA even in the early stages of apoptosis because the monoclonal antibody to single-stranded DNA (ssDNA) does not react with ssDNA ends present in necrotic cells [27] .
Briefly, tissue from three larvae for each condition were used to reduce the individual effect and incubated for 6 h at 4˚C and for 20 min at 37˚C in RPMI 1640 medium +10% heat-inactivated horse serum +5% heat-inactivated fetal bovine serum and 0.25% trypsin (Invitrogen 2011). Then, the cell suspension was filtered through a nylon cell sieve (100 µm pores) (BD Biosciences, Mississauga, Ontario, Canada) and about 8000 cells per well were placed in a 96 wells plate and harvested for 5 min at 300 g. Culture medium was removed and the plate was incubated for 30 min at RT in 200 µl of fixative solution (80% (v/v) methanol in PBS) and then dried overnight at RT. A second plate containing 100 µl of positive control was placed overnight at 37˚C. The following morning, plates containing experimental cells as well as control cells were incubated for 10 min in 50 µl of formamide at RT, followed by incubations of 20 min at 75˚C and 5 min at 4˚C. Then, 100 µl of S1 nuclease (Invitrogen, Burlington, Ontario, Canada) were added to a negative control well and incubated for 30 min at 37˚C. From this step on, the two plates are treated in the same manner: three washes of 5 min with PBS and 1 h incubation at 37˚C in 3% (w/v) non-fat dry milk in distilled water. Milk was removed and the plates were incubated in 100 µl of antibody solution for 30 min at RT. The antibody solution is a mix of primary and HRP-coupled secondary antibodies; just one incubation period is needed and the received signal is specific [27] . Three washes of 5 min were completed with Washing Solution from the kit, and the ABTS (2, 2’-AZINO-bis [3-éthylbenziazoline-6-sulfonic acid]) was added. A specific signal was detect by absorbance at 405 nm wavelength, on a microplate reader (Fisher Scientific, Ottawa, Ontario, Canada) after about 30 min of incubation.
2.5. Detection of Protein Markers of Apoptosis
Total proteins were extracted from a blend of three larvae according to the total extraction method from the Nuclear Extract Kit (Active Motif, Carlsbad, CA). The protein concentration was determined by using the BCA protein assay kit (Pierce Biotechnology inc., Rockford, IL). Expression of Bax, Bcl-2, p53 and PARP-1 proteins was quantified by Western blot as already reported [26] . Briefly, equal amounts of proteins were loaded on a 10% SDS-polyacrylamide gel, and an electrophoretic separation was run for 5 h at 120 V. The gels were transferred onto PVDF membranes (0.2 µm pores, Bio-Rad, Hercules, CA) then the blots were blocked for 1 h at room temperature (RT) in 5% non-fat dry milk and incubated in the primary antibody solution for 24 h. Polyclonal primary antibodies used were: rabbit anti-Bax (1:200, Delta Biolabs, Gilroy, CA), rabbit anti-Bcl-2 (1:100, Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-p53 (1:200, Santa Cruz Biotechnology, Santa Cruz, CA) and rabbit anti-full-length PARP-1 (1:50, Santa Cruz Biotechnology, Santa Cruz, CA), all diluted in TBS-T with 0.5 g BSA and 25 mg sodium azide. Blots were washed and incubated with HRP-conjugated polyclonal secondary antibody goat anti-rabbit (1:10,000) diluted in 5% non-fat dry milk for 2 h at RT and developed with an enhanced chemiluminescence substrate solution [28] . Images were acquired with an AlphaEase FC imaging system (Alpha Innotech, San Leandro, CA) and analysed with AlphaEase FC Software (Alpha Innotech).
2.6. Statistical Analysis
We used a general linear mixed modelling approach to compare the effects of light quantity and quality on yellow perch larval apoptosis and survival. For each variable studied, we built a mixed model containing all variables that could predict the response variable: light quality and intensity (fixed effects), interaction between light quality and intensity (fixed effect) and block effect (random effect). Survival data (%) were arcsin transformed before their integration to the models. We used an information—theoretic approach to compare and rank the models [29] . The Akaike Information Criterion (AIC) is selecting the best model to explain variation with the fewest variables. All models are ranked using , where AICi is the AIC value for a given model and AICmin is the smallest AIC value for the compared models. When
, where AICi is the AIC value for a given model and AICmin is the smallest AIC value for the compared models. When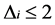 , a model has an important support, when
, a model has an important support, when  the support is considerable and when
the support is considerable and when  the model has less support. We used the AIC corrected for small size samples (AICc) for model selection because the ratio of the sample size and the number of parameters in the model is lower than 40 [29] . Model selection was based on the procedure suggested by [30] . Briefly, we first compared the fit of two models containing all the fixed factors but differing by the presence of the random term. Restricted maximum likelihood estimation (REML) methods were used at this stage to determine if the random factor should be included in the model [30] . Second, we tested the contribution of the fixed factors to the fit of the model, by comparing models differing only in their fixed part. Maximum likelihood estimation (ML) methods were used at this stage [30] . At the end of the process, the selected model was refitted using REML. If the selected model included a random factor, we calculated the percent of variation explained by each model by squaring the Pearson correlation between observed values and values predicted by the full model [31] . Otherwise, adjusted R² were calculated by refitting the data with a linear model. Statistical analysis were made with the functions lme(nlme), gls(nlme) and lm(stat) in the R© language (Version 2.12.1, 2010).
the model has less support. We used the AIC corrected for small size samples (AICc) for model selection because the ratio of the sample size and the number of parameters in the model is lower than 40 [29] . Model selection was based on the procedure suggested by [30] . Briefly, we first compared the fit of two models containing all the fixed factors but differing by the presence of the random term. Restricted maximum likelihood estimation (REML) methods were used at this stage to determine if the random factor should be included in the model [30] . Second, we tested the contribution of the fixed factors to the fit of the model, by comparing models differing only in their fixed part. Maximum likelihood estimation (ML) methods were used at this stage [30] . At the end of the process, the selected model was refitted using REML. If the selected model included a random factor, we calculated the percent of variation explained by each model by squaring the Pearson correlation between observed values and values predicted by the full model [31] . Otherwise, adjusted R² were calculated by refitting the data with a linear model. Statistical analysis were made with the functions lme(nlme), gls(nlme) and lm(stat) in the R© language (Version 2.12.1, 2010).
3. Results
3.1. Cumulative UVR Exposure
Based on the incident irradiance measured above the filters, we calculated that the cumulative UVR exposures received for the 313 nm (UV-B) and 340 nm (UV-A) wavelengths were of 0.67 and 3.35 W∙m−2∙nm−1, respectively. The UVA/UVB ratio [calculated as UV-A/(UV-A + UV-B)] was of 0.83. Therefore, UVR daily exposures (Table 1) were very similar to those measured in our previous experiments [7] .
3.2. Effects of UVR Irradiance and Spectral Composition
The random factor (block) was not included in any of the models since, for all the variables measured; the AICc suggested that its inclusion did not improve their fit.
3.3. Survival Rate
None of the treatments, quality and/or intensity affected larvae survival after two days of exposure (Figure 1).
According to the Δi, the best model to explain the yellow perch larvae survival rate includes only the light intensity (model D in Table 2(a)). Besides being the best one, this model explains virtually no variation in larvae sur

Table 1 .Cumulative exposures and UV-A/UV-B ratios received by each treatment for the two experiments as estimated by continuous irradiance measurements (GUV-2545/2456) and calculated transmittance of the different filters. Exp = experiment; Light quantity: high = no screen, medium = one screen, low = two screens.

Table 2. Set of candidate models to explain survival rate, p53 expression, Bax/Bcl-2 ratio, PARP-1 expression and DNA fragmentation of Yellow perch larvae. The selection of the model (in bold) was based on the Akaike information criterion corrected for small samples (AICc; see text for more details). All the comparisons are based on maximum likelihood estimates. (a) survival rate; (b) p53 expression; (c) Bax/Bcl-2 ratio; (d) PARP-1 expression; (e) DNA fragmentation. µ = intercept, x1 = spectral quality, x2 = light intensity.
vival (F2,15 = 0.46, P = 0.630,  < 0.01), suggesting that neither spectral quality nor intensity of UVR had a significant effect on the mortality of larvae during the two first days of exposure (Figure 1).
< 0.01), suggesting that neither spectral quality nor intensity of UVR had a significant effect on the mortality of larvae during the two first days of exposure (Figure 1).
3.4. Specific Detection of Apoptotic DNA
We initially measured specific DNA fragmentation, detected by formamide-induced denaturation of specific apoptotic ssDNA, as already reported [26] [27] [32] [33] . This assay is highly specific for apoptotic cells and does not demonstrate reactivity with necrotic cells or cells containing DNA breaks in the absence of apoptosis [27] . Specific apoptotic DNA fragmentation was lowest under visible light + UV-A + UV-B and under visible light + UV-A and higher under visible light treatment, whatever the light intensity (Figure 2). The best model to explain the apoptotic DNA fragmentation in Yellow perch larvae exposed to different light treatments was the one including only light spectral quality (model C in Table 2(e)). This model explains a considerable fraction of the variation in DNA fragmentation (F2,15 = 13.55, P < 0.001,  = 0.596).
= 0.596).
3.5. Expression Protein Markers of Apoptosis: Bax/Bcl-2, P53 and PARP-1
We studied the expression of p53 (Figure 3), a tumor suppressor protein known to play an important role in evoking apoptosis, notably by encouraging the transcription of several pro-apoptotic genes such as Bax [34] . The best model to explain the p53 expression included only spectral composition (model C in Table 2(b)) and explained virtually no variation in p53 protein expression (F2,15 = 0.48, P = 0.630,  < 0.01). The model including only light intensity (model D in Table 2(b)) had virtually the same fit (Δi < 2; F2,15 = 0.12, P = 0.890,
< 0.01). The model including only light intensity (model D in Table 2(b)) had virtually the same fit (Δi < 2; F2,15 = 0.12, P = 0.890, 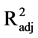 < 0.01). Therefore, neither spectral quality nor intensity affected p53 expression during the two days of exposure (Figure 3).
< 0.01). Therefore, neither spectral quality nor intensity affected p53 expression during the two days of exposure (Figure 3).
The variation in Bax/Bcl-2 ratio, reported to be correlated with apoptosis [35] , was best modeled by two models with similar fit (model C in Table 2(c)). The models included either spectral quality or light quantity and explained virtually no variation in the ratio of Bax and Bcl-2 (F2,15 = 1.29, P = 0.303,  < 0.01, and F2,15 = 0.57, P = 0.580,
< 0.01, and F2,15 = 0.57, P = 0.580,  < 0.01, respectively). Therefore, this variable is not affected by spectral quality or intensity of UVR after two days of solar exposure (Figure 4).
< 0.01, respectively). Therefore, this variable is not affected by spectral quality or intensity of UVR after two days of solar exposure (Figure 4).
We also studied PARP-1 protein expression, a major player in the prevention of apoptosis. The best model to explain PARP-1 expression included only spectral composition (model C in Table 2(b)) and explained virtually no variation in the expression of this protein (F2,15 = 0.889, P = 0.43,  < 0.01). Therefore, neither spectral quality nor intensity affected PARP-1 expression during the two days of exposure (Figure 5).
< 0.01). Therefore, neither spectral quality nor intensity affected PARP-1 expression during the two days of exposure (Figure 5).

Figure 3. Effect of UVR quality and intensity on p53 protein expression. Dark grey: Whirlpak® (visible + UV-A + UV-B), light grey: Mylar-d® (visible + UV-A), and white: J-Roll® (visible). None of the treatments influences p53 protein expression after two days of UVR exposure.

Figure 4. Effect of UVR quality and intensity on the Bax/Bcl-2 ratio of protein expression. Dark grey: Whirlpak® (visible + UV-A + UV-B), light grey: Mylar-d® (visible + UV-A), and white: J-Roll® (visible). None of the treatments influences Bax/Bcl-2 protein ratio after two days of UVR exposure.

Figure 5. Effect of UVR quality and intensity on full-length PARP-1 protein expression. Dark grey: Whirlpak® (visible + UV-A + UV-B), light grey: Mylar-d® (visible + UV-A), and white: J-Roll® (visible). None of the treatments modulate full-length PARP-1 expression.
4. Discussion
Our results show that larvae mortality did not differ among different levels of spectral quality and light intensity in field conditions, thus suggesting that UVR have no effects on larvae survival after two days of solar exposure. Our data also show that the UV-A and UV-B parts of the UVR can reduce the apoptosis rate in still-living yellow perch larvae, as demonstrated by specific apoptotic DNA fragmentation analysis, while the expression of apoptotic protein markers p53, PARP-1 and the Bax/Bcl-2 ratio did not show any clear response to UVR.
Whereas it might be argued that these results are due to a reduced sample size and, in turn, to low statistical power, this argument cannot be applied to the result of specific apoptotic DNA fragmentation, indicating an important decrease of apoptosis in samples exposed to UV-A + visible or UV-B + UV-A + visible. Then we used a specific ELISA test based on the selective denaturation of DNA by formamide in apoptotic cells, and detection of denatured DNA with monoclonal antibody to single-stranded DNA. Formamide is a gentle agent that denatures DNA in apoptotic cells but not in necrotic cells or in cells with DNA breaks in the absence of apoptosis [26] [27] [32] [33] . Importantly, the sensitivity of DNA in apoptotic cells to formamide is not related to DNA breaks, but rather reflects changes in chromatin associated with apoptosis, such as chromatin condensation and digestion of protein stabilizing DNA. By suggesting a reduction of apoptosis in still-living larvae due to UVR exposure, our results on this specific apoptotic indicator are rather intriguing, but are conservative since this response is clear despite the low statistical power.
Under conditions similar to ours (i.e., same experimental setup, location and time of the year) but after a longer UVR exposure (i.e., seven days), [23] showed that UV-A and UV-B can have a strong negative effect on Yellow perch larvae survival. Other studies showed that UVR reduces fish larvae survival [22] [36] , but the mortality rate was measured following four to ten days of UVR exposure. Thus, it may be possible that two days exposure, as in this study, is not sufficient to affect the larvae survival. However, DNA fragmentation may be already modulated inside the cell of living larvae, as our present results demonstrate.
We also investigated the protein expression of several markers of apoptotic pathways and found that the exposure to natural UVR for two days did not modulate the expression of p53, Bax/Bcl-2 ratio and full-length PARP-1 in still-living yellow perch larvae. These results are compatible with our finding showing decreased levels of specific apoptotic DNA fragmentation in the same field conditions, and altogether they sustain a differential sensitivity of yellow perch larvae during development, as similarly reported for coral larvae [37] .
Bcl-2 is a key member of the anti-apoptotic Bcl-2 family and is a potent inhibitor of apoptotic cell death [35] . Bcl-2 and Bcl-xL preserve mitochondrial integrity and prevent the subsequent release of apoptogenic molecules such as cytochrome c [38] . On the other hand, Bax is also a member of the Bcl-2 protein family. It accelerates the rate of apoptosis by a contribution to the permeabilization of the outer mitochondrial membrane, either by forming channels by themselves or by interacting with components of the outer mitochondrial membrane pore [39] . A high Bax/Bcl-2 ratio indicates cellular improvement to apoptosis, as already reported [40] . Our data do not support the hypothesis that Bax/Bcl-2 protein ratio is modulated following exposure to UVR. We also studied the expression of p53, a tumor suppressor protein known to play an important role in evoking apoptosis, notably by encouraging the transcription of several pro-apoptotic genes such as Bax [34] and the expression of full-length PARP-1, a protein known to participate in the repair of damaged DNA [41] . Our results do not support the view that the expression of the full-length PARP-1 protein is modulated in presence of UV-A and UV-A + UV-B, for a short term exposition. Indeed, when the DNA damages are limited, PARP-1 activates DNA reparation and is released once the DNA is completely repaired, without being cleaved (see for an extensive review [42] ), as sustained in our experiments. As the two treatments, visible light +UV-A and visible light + UV-A + UV-B, contain the UV-A part of the solar spectrum; it is also possible that UV-A activates the enzyme CPD photolyase. This enzyme binds to a CPD in DNA strain; it is directly activated by the 350 - 450 nm wavelengths and breaks the links between the two pyrimidines, without damaging the cell [19] . Although we performed specific experiments to detect a possible modulation of clived PARP-1 and clived caspase-3 in field condition, we could not present any results due to the poor specificity of various anti-clived PARP-1 and anti-caspase-3 antibodies tested to perform western blot techniques, on our preparation of fish larvae total protein extract.
In our experimental paradigm, three out of four proxies of apoptosis did not respond to variations in light quality or quantity. Whereas this may suggest that the exposure time was too short to determine any effect on yellow perch larvae, there are clear indications that this was not the case. Apoptosis was yet onset, as showed by variation in specific apoptotic DNA fragmentation among the treatments. Indeed, this specific apoptotic marker clearly responded to changes in spectral quality. These data are statistically significant despite the low sample size and short-term exposure (both increasing the risk of type-II error), suggesting that the signal we detected for DNA fragmentation was strong enough and thus should not be underestimated, even though it goes against our initial hypotheses.
Because UVR is known to induce apoptosis in culture cells [43] -[45] , we were expecting that UV-A and UV-B would increase specific DNA fragmentation and the expression of the other apoptotic parameters. In contrast, in our field conditions, we detected a decrease of apoptotic DNA fragmentation and no significant variations of p53, PARP-1 expression and Bax/Bcl-2 ratio. These apoptotic markers are considered as indicators of the intrinsic pathway of apoptosis and usually, their modulation reflect an on-going apoptotic process [7] [26] [33] [46] . This apparent contradiction may be explained by UVR inactivating the intrinsic or mitochondrial apoptotic pathway initiated by UVR-induced damage to DNA [10] . Our hypothesis is supported by several studies revealing that different apoptotic pathways may be initiated by UVR exposure [9] [21] [47] [48] . As UVR may initiate the apoptotic pathway through the intrinsic and the extrinsic, as well as the mitogen-activated protein kinases or the ROS pathways, it is possible that one of those is induced faster than the intrinsic pathway. Actually, many data measuring UV-induced apoptosis have used experimental protocols performed several hours after the UV incubation. This delay allows the cell to enter in the apoptotic cascade and to produce proand anti-apoptotic proteins [44] [49] [50] . On the other hand, we performed our cellular experiments immediately after harvesting still-living larvae exposed to two days of natural UVR.
Alternatively, the existence of a time window of low sensitivity to UV-A and UV-B may correspond to a specific larvae developmental stage where basal level of apoptosis should be moderated in favour of mitotic pathways. Indeed, it has been recently reported that coral larvae exhibit low sensitivity to natural levels of UVR during early development stages [37] reflected in few transcriptional changes, while later in coral larvae development, these authors report that natural UVR induces genes related with stress response, the endoplasmic reticulum, Ca2+ homeostasis, development and apoptosis. More specifically, they also observed a complex response combining upand down-regulation of apoptosis-inducing factors as well as inhibitors [37] .
Our results clearly show that UV-A and UV-B reduce apoptotic DNA fragmentation in still-living Yellow perch larvae exposed to natural light. However, several studies on the effects of UVR in aquatic organisms showed that UVR, mostly UV-B, are responsible for the formation of CPD in DNA, eventually leading to the fragmentation of the DNA [7] [51] [52] . Other studies also showed that UVR activates the apoptosis pathway in severe stress conditions, such as ultraviolet light at 10-100 mJ/cm2 [50] . It should be noted that the majority of studies performed on UV and aquatic organisms measure dose-dependent increases in the level and/or in the activation of stress markers, consistent with their role in mediating cellular response to stress and suggesting a function in larvae survival [20] [21] [53] . In our study, under natural UVR in field condition, the specific apoptotic DNA fragmentation in the visible light condition (Figure 2) is higher than in treatments where UV-A is present. This may be explained by the fact that CPD photolyase, activated by UV-A, reduces the DNA damages and may decrease the apoptotic cascades in Yellow perch larvae, suggesting that UVR in field condition may reduce apoptosis in organism where the apoptotic pathways are important for their morphogenesis.
5. Conclusion
In conclusion, this is one of the very few original experimental studies on apoptosis parameters in fish larvae conducted under natural light. Despite the short term exposure to UVR, our results clearly show that these wavelengths can affect specific apoptotic DNA fragmentation, an hallmark of the apoptotic process. By showing that specific DNA fragmentation can be decreased rather than increased by exposure to UVR, our results open new ways to the understanding on the effects of this stressor on the early development of living organisms in natural exposure conditions. Understanding whether developing organisms are affected by UVR (increased stress or impaired morphogenesis) or they can set on their metabolic machinery to reduce UVR damages (DNA photorepair induced by UV-A), may be a key step to predict the outcome of UVR exposure in living organisms. It is now important to more explicitly address these questions by measuring simultaneously the photorepair and the apoptotic response in organisms exposed to UVR during the key steps of their development and also analyse more specific morphogenesis in yellow perch larvae.
Acknowledgements
This study was funded by grants from the Natural Sciences and Engineering Research Council of Canada to P. M. (No. 15851) and to M.-G. M. (No. 170571) and from the Canada Research Chair Program to P. M. We thank Dr. J.-J. Frenette, C. Fournier, D. Gadbois-Côté, G. Pepin, F. Longpré, J. Carange and J. Bournival for their assistance.
NOTES
*Corresponding author.