1. Introduction
Candida albicans are classified as commensal fungi that inhabit the human gastrointestinal tract. It grows as round cells in smooth, white colonies. Additionally, C. albicans can cause oral and vaginal infections as well as systemic diseases [1]. Vaginal candidosis (VC) is one of the most common infections that afflicts women of reproductive age [2,3]. Approximately 75% of women will experience at least one episode of VC during their lives [3,4]. C. albicans possesses several virulence factors that are involved in hyphae formation [5], phenotype switching [6-8], cell adhesion [9-12], and extracellular production of hydrolytic enzymes [13,14]. Secreted aspartyl proteases (Saps) are enzymes that are secreted by C. albicans and are coded for by the SAP gene family (SAP1-SAP10) [15]. The proteolytic activity of the Sap proteins is involved in the degradation of the host’s barriers during infection [16], immune response evasion [17], and adhesion to the host’s cells [18]. This family of proteases can be differentially expressed and regulated under a variety of growth conditions in the laboratory [19,20], such as during experimental infections using reconstituted human oral epithelium (RHE) [21], or in vivo [22].
Different SAP genes appear to be essential for mucosal (SAP1-SAP3) [23,24], and systemic infections (SAP4- SAP6) [25]. The expression and importance of SAP1, SAP2, SAP3 during murine vaginal candidiasis were also demonstrated by using RT-PCR and SAP-deficient mutants [25-27]. It is not clear whether the murine model is representative of proteinase expression during human vaginal infection.
The purpose of this work was to use a reconstituted human vaginal epithelium (RHVE) model to determine the frequency and expression of the ten SAP genes in C. albicans strains that had been isolated from women with VC.
2. Material and Methods
2.1. Patients and Samples
Procedures followed in this study were in accordance with the Ethical Committee of each hospital.
This study included 264 women who presented with vulvovaginitis-associated symptoms, such as burning, itching, dysuria, and curd-like discharge; these women visited gynecologic services at public hospitals located in the State of Mexico, Mexico. The age range of the women in this study was between 18 and 57 years. Patients presenting with cervical cancer, pregnancy, or who had undergone antibiotic or antimycotic treatments within the last 30 days were excluded from the study. After obtaining informed consent from each patient, two samples were taken from the vaginal cavity using sterile cotton swabs. One of these samples was used to confirm the presence of pseudohyphae or hyphae by direct examination under a microscope. The second sample was used to inoculate brain hearth infusion (BHI) culture media (BD Bioxon, Cuatitlán Izcalli, State of Mexico, Mexico), and the cultures were then incubated at 37˚C for 24 hours. After 24 hours, the cultures were plated on Sabouraud Agar (BD Bioxon, Cuatitlan Izcalli, State of Mexico, Mexico) containing 50 µg/ml chloramphenicol and were incubated at 37˚C for 72 hours.
2.2. Identification of C. albicans
Samples were collected from the pure cultures grown in Sabouraud Agar and were identified by colony and microscopic morphologies and using a germ tube test in BHI that was supplemented with 10% horse serum and the API 20 C AUX system (BioMerieux, Durham, NC, USA). The C. albicans ATCC32354 strain was used as a positive control.
C. albicans was also identified using PCR by amplifying the internal transcribed spacers (ITS1 and ITS2) from the rRNA gene [28]. C. albicans genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA). PCR amplification was performed in a Corbette Research Thermocycler using PuReTaq Ready-To-Go PCR Beads (GE Healthcare, Piscataway, New Jersey, USA). The amplicons were stained with ethidium bromide after electrophoresis in a 2% agarose gel and were visualized with UV illumination using a GEL LOGIC 100 (KODAK). The C. albicans ATCC32354 strain was used as a positive control.
2.3. PCR Amplification of C. albicans SAP Genes
The primers and amplification conditions for SAP1 were described by Hube et al. [29], SAP2 by Wright et al. [30], SAP3 by White et al. [31], SAP4-SAP6 and SAP8 by Naglik et al. [22], SAP7 by Monod et al. [32], and SAP9 and SAP10 by Naglik et al. [33] (Table 1).
Table 1. Primers used in PCR and real time PCR assays.

2.4. RHVE Inoculation with C. albicans
Both C. albicans strains tested positive for the expression of all of the SAP genes, and the control strains were grown as described by Schaller et al. [16,21]. The C. albicans ATCC32354 strain was used as a positive control. The following strains from our collection, which are each missing one SAP gene, were used as negative controls: C. albicans 4, C. albicans 7, C. albicans 22, Staphylococcus epidermidis ATCC35984, C. albicans 27, C. albicans 50, and C. albicans 28. A total of 2 × 106 C. albicans cells suspended in 50 µl of PBS were inoculated onto the surface of the RHVE A431 (SkinEthic Laboratory, Nice, France) and were incubated at 37˚C for 72 hours with 5% CO2 and saturated humidity. The maintenance media was changed every 24 hours.
2.5. C. albicans RNA Purification and Reverse Transcription
C. albicans cells were harvested from the RHVE, suspended in 200 µl of Y1 buffer containing lyticase (50 U/107 cells), and incubated at 30˚C for 20 minutes using gentle shaking to facilitate the formation of spheroplasts. The extraction and purification of the total RNA was performed using an RNeasy MiniKit (Qiagen, Hilden, Germany). The RNA concentration and purity were determined using a Nanodrop 2000 spectrophotometer. To obtain cDNA, a QuantiTec Reverse transcription kit (Qiagen) was used according to the manufacturer’s instructions.
2.6. Real-Time PCR Amplification of the SAP Genes
The SAP primers that were used for the endpoint PCR were also used for the real-time PCR assay (Table 1). The Rotor-Gene SYBR Green PCR kit (Qiagen) was used for the real-time PCR experiments. A final volume of 25 µl was used for each reaction and contained 12.5 µl of SYBR Green Master Mix, 1 µl of forward primer (1 µm), 1 µl of reverse primer (1 µm), 2 µl of cDNA (20 ng), and 8.5 µl of RNase-free water. The amplification conditions were 95˚C for 5 minutes followed by 95˚C for 5 seconds with an annealing/extension combination step at 60˚C for 10 seconds for 40 cycles. The same positive and negative controls that were used for the endpoint PCR experiments were also used for the real-time PCR assay.
2.7. Statistical Analysis
The frequency of the SAP family genes among the C. albicans strains isolated from women within the age ranges of 18 - 29 (n = 13), 29 - 39 (n = 14), 40 - 49 (n = 11), and 50 - 57 (n = 12) years were analyzed using the c2 test with a significance threshold of P < 0.005.
3. Results
3.1. C. albicans Positive Samples
Infection with C. albicans was determined using microbiologic criteria and by PCR. The ITS were amplified from 18.9% (n = 50) of the vaginal samples (Figure 1).
3.2. Detection and Association of SAP Genotypes in C. albicans Strains
The SAP4, SAP5, SAP6, SAP2, SAP8, SAP1, and SAP10 genes were detected at a higher frequency in C. albicans strains than were the SAP9, SAP3, and SAP7 genes (Table 2). There were no statistically significant differences among the SAP genotype frequencies in the C. albicans strains isolated from women in the age ranges of 18 - 29 (n = 13), 29 - 39 (n = 14), 40 - 49 (n = 11), and 50 - 57 (n = 12) years (threshold of P < 0.005).
Sixteen different association patterns of SAP genes were found among these strains (Table 3). Twenty strains carried all of the studied SAP genes, and ten of the strains had only nine of the SAP genes (Table 3).

Figure 1. Candida albicans identification by conventional PCR. (ITS, 273 bp amplicon): Lanes 1 - 3 and 5, C. albicans isolates; Lane 4, MWM 50-bp ladder; Lane 6, positive control C. albicans ATCC32354; Lane 7, reagent control mixture without DNA.
Table 2. Frequency of the SAP genes and their expression in Candida albicans strains.
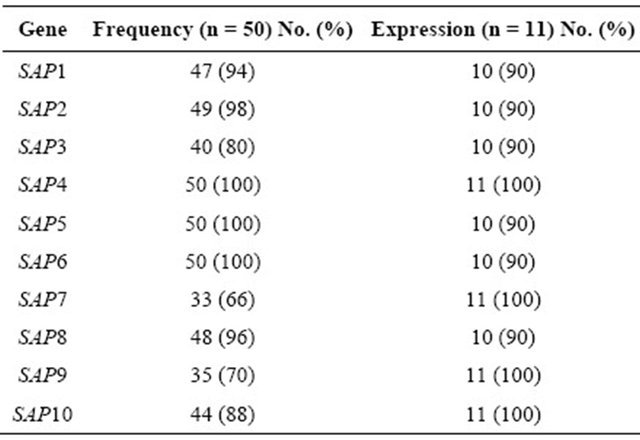
Table 3. Detected SAP genotypes in Candida albicans strains.

3.3. SAP Gene Expression Determined by Real-Time PCR
To determine the SAP virulence markers, 11 clinical strains carrying all of the SAP genes (Pattern 1, Table 3) were inoculated onto RHVE and were then analyzed using real-time PCR. The Tm was recorded for each realtime assay to differentiate specific from non-specific PCR products (data not shown). Constitutive rRNA expression was used as a control (data not shown). All of the SAP genes were expressed. The genes SAP4, SAP7, SAP9, and SAP10 were expressed in 100% of the isolated C. albicans strains, whereas SAP1, SAP2, SAP3, SAP5, SAP6, and SAP8 were expressed in 90% of the strains (Table 2, Figure 2).
4. Discussion
Candida albicans was identified in vaginal samples of 18.9% (n = 50) of the women in this study. This result is in agreement with previously reported frequencies for VC, which occurs in 20% - 25% of infectious vaginitis cases, which is just below the 40% - 50% of cases that develop bacterial vaginitis [4]. Among Mexican women, VC is the ninth most common cause of disease (incidence of 529 in 100,000 women; SUIVE/DGE/Ministry of Health/United Mexican States-2008). Candidosis is
not common during puberty; however, by 25 years of age, 50% of women have had at least one clinically diagnosed VC episode [34,35]. C. albicans causes 85% - 95% of VC cases [4,36]. External factors that predispose patients to VC are pregnancy, the use of oral contraceptives, diabetes mellitus, and antibiotics usage [3,37].
The high frequency of SAP genes (Table 2) and the high number of association patterns in C. albicans strains (Table 3) suggest that multiple Sap expression profiles exist during VC pathogenesis. For example, the SAP4- SAP6 genes, which were present in all of the analyzed C. albicans strains (Table2), are associated with hyphae formation. In addition to being involved in cell adhesion, hyphae can penetrate individual cells or intercellular spaces [38]. The Sap2 protein, which was detected in 98% of the studied strains (Table2), can degrade many human proteins, including those that protect the mucosal surfaces, such as mucine [39,40] and secretory immunoglobulin A (sIgA) [41], and molecules from the extracellular matrix, such as keratin, collagen, and vimentin [42- 44]. The ability of Sap2 to degrade these proteins may facilitate the dissemination of C. albicans throughout the circulatory system [45].
In this study the genes SAP4, SAP7, SAP9, and SAP10 were expressed in 100% of the isolated C. albicans strains, whereas SAP1, SAP2, SAP3, SAP5, SAP6, and SAP8 were expressed in 90% of the strains (Table 2). Similar data were described by Schaller et al. [46], who used an in vitro candidosis RHVE model. In their study, the genes that were most frequently expressed during the late infection stage were SAP1, SAP2, SAP4, SAP7, SAP9, and SAP10, whereas in studies performed on vaginal infections in vivo, the most frequently expressed genes were SAP1, SAP3, and SAP6-SAP8 [22]. In vulvovaginal candidosis (VVC) and recurrent vulvovaginal candidosis (RVVC), the expression of SAP2 and SAP4- SAP7 [47] was reported. It has been suggested that the SAP4-SAP6 genes, which are frequently expressed in vaginal infection samples, play an important role in the evasion of the immune response during the active infection stage, which partially protects C. albicans from macrophage phagocytosis [17]. It has also been reported that the expression of the SAP4-SAP6 genes is associated with hyphae development in vitro [48], which suggests that Sap4-Sap6 production in the vaginal lumen enables C. albicans to attach to the vaginal mucosa.
Results presented in this study showed that all of the SAP genes were expressed in the RHVE, suggesting that the Sap proteins play an important role in the pathogenesis of infection.
5. Acknowledgements
This work was supported by the grant PAPIME PE200209 from Universidad Nacional Autónoma de México.
NOTES
#Corresponding author.