A Modified Selective Medium Containing Benzalkonium Chloride (BKC) for the Isolation of Pseudomonas aeruginosafrom Raw Milk ()
1. Introduction
Psychrotrophic bacteria, of which Pseudomonas is an important genus, are considered the most important pathogens contributing to milk spoilage through the production of lipolytic and proteolytic enzymes [1,2]. Many of these enzymes are not inactivated by pasteurizing at 72˚C for 15 s or by Ultra-High Temperature (UHT) treatment. The residual activities of these enzymes can reduce the organoleptic quality and shelf life of processed milk products [3]. Gram-negative bacteria usually account for more than 90% of the microbial population in cold raw milk and among these bacteria Pseudomonas are the predominant microflora [4].
Although several selective media for the detection of Pseudomonas species are produced commercially, they lack specificity [1]. Conventional methods for the detection of P. aeruginosa use cetrimide as a selective agent. Cetrimide Agar and Pseudosel Agar (BBL) are devised to serve as selective media for the isolation and identification of Pseudomonas aeruginosa while supposedly inhibiting the growth of other microorganisms [5,6]. The 0.03% Cetrimide Agar is more selective for growth of P. aeruginosa than is Pseudosel Agar; however, certain bacteria other than P. aeruginosa also grow on the former medium. Although Pseudosel Agar is not a highly selective medium for P. aeruginosa, it is preferable to Technicolor Agar for detection of the pyocyanin and pyorubin pigments produced by P. aeruginosa [5]. Therefore, the performance of available culture method is of great importance. Most frequently, Cetrimide Broth or Cetrimide Agar or MacConkey Agar are used [7]. Although Cetrimide Agar is strongly selective for P. aeruginosa, some strains of Gram-negative bacilli (especially Klebsiella spp. and Providencia) can grow on it [8]. The potent inhibitory substances in these media not only suppress the competing bacterial flora but may also exert an adverse effect on the targeted bacterium [9].
There is an increasing concern that the use of biocides such as benzalkonium chloride (BKC) [10]. BKC has a broad-spectrum antimicrobial activity [10,11]. Most of P. aeruginosa strains resistant and adapted to BKC also results in higher resistance to amphoteric disinfectants [12].
The aim of the current study was to develop an improved selective Cetrimide Agar for the isolation of Pseudomonas aeruginosa from raw milk. BKC was chosen as a selective agent for the development of a modified selective Agar, because BKC has an important effect on variety of Pseudomoas species and other pathogens which are able to grow on Cetrimide Agar and does not have an inhibitory effect on Pseudomonas aeruginosa. In this study, most acceptable concentration of BKC was tried to detect to make Cetrimide Agar base more selective for Pseudomonas aeruginosa.
2. Materials and Methods
2.1. Microorganisms and Media
Pseudomonas aeruginosa ATCC 29212 was used as a reference strain for this study. Other bacterial strains were isolated from raw milk samples collected from several dairy plants in Turkey. Cetrimide Agar (CA) used for isolation was prepared from dehydrated powders in accordance with manufacturer’s instructions (Merck, Darmstadt, Germany). In order to prepare modified CA (mCA) plates, CA at 50˚C was supplemented with BKC which was used as a selective agent to improve selectivity of CA. In addition to CA medium, Tryptic Soy Agar (TSA, Merck, Darmstadt, Germany) was used as the control medium.
2.2. Isolation and Identification
Raw milk samples that were carried to laboratory at 4˚C were plated on Cetrimide Agar by spread plate technique and the plates were incubated at 28˚C for 72 h. The colonies isolated on these plates were identified using Microbact 24E Gram negative Identification System (Oxoid, Cambridge, UK). Stock cultures were maintained at –21˚C in Tryptic Soy Broth (TSB, Merck, Darmstadt, Germany) containing 20% (v/v) glycerol.
2.3. Determination of Minimum Inhibitory Concentrations (MICs) for BKC
MICs for BKC were determined by Agar dilution method. MICs were performed with various concentrations of BKC (50, 100, 150, 200, 250, 300, 350, 400, 450, 500, 550, 600 µg/mL) in Cetrimide Agar. P. aeruginosa and other Gram (-) bacteria were grown at 18 h on TSB and 2 µl aliquot of each strain was inoculated onto each of the Agar plates, then the plates were incubated at 37˚C overnight. The lowest concentration preventing growth of cultures was taken to be the MIC [13]. This experiment was performed in duplicate.
3. Results and Discussion
3.1. Isolation and Identification
Many selective agents such as cetrimide, cephaloridine, fucidin nalidixic acid, phenanthroline, Irgasan have been used to obtain selective media for isolation and identification of Pseudomonas aeruginosa. Although several selective media for P. aeruginosa have been described, in fact none of them is absolutely specific [13]. Nowadays, Cetrimide Agar is commonly used for isolation and identification of Pseudomonas aeruginosa; however studies indicate that some strains of Gram-negative bacilli can grow on it [8].
A total of 14 raw milk samples were examined for isolation of P. aeruginosa and other Gram (-) bacteria using Cetrimide Agar and a total of 55 isolates were obtained and identified. Pseudomonas spp. accounted for 85.5% of the total isolates and among these isolates P. aeruginosa and P. fluorescens accounted for 34.6% and 50.9%, respectively.
The main psychrotrophic microflora encountered in raw milk are Gram-negative rods composed mainly of psychrotrophic species of Pseudomonas, Achromobacter, Aeromonas, Serratia, Alcaligenes, Chromobacterium, Flavobacterium and Enterobacter. In milk, Pseudomonas spp. comprise at least 50% of the total bacteria [3]. In the current study, Pseudomonas was the most predominant genus, and P. fluorescens was the most predominant species isolated from raw milk samples. In addition to these bacteria, Acinetobacter baumannii, Enterobacter intermedium, Enterobacter agglomerans and Escherichia coli accounted for 7.2%, 3.6%, 1.8% and 1.8% of the 55 isolates, respectively (Table 1). It is known that the predominant flora of the raw milk is P. fluorescens. Ternström et al. [14] observed that P. fluorescens was present in 84% of the raw milk samples. Similarly, Gennarl and Dragotto [15] showed that P. fluorescens accounted for 55.6% of the all bacterial isolates in raw milk. Hence, there is a need to inhibit these other Gram (-) bacteria especially P. fluorescens, so as to enhance selectivity of CA for P. aeruginosa.
3.2. MICs for BKC
Benzalkonium chloride is a nitrogen-based quaternary
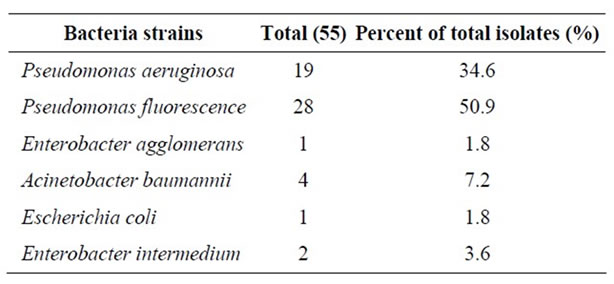
Table 1. Total number of Pseudomonas aeruginosa and other Gram-negative bacteria isolated from raw milk samples on Cetrimide Agar.
ammonium compound (QAC) demonstrating broad-spectrum antimicrobial activity [12]. It is well known that Gram-negative bacteria are less susceptible to QACs than Gram-positive bacteria, and Pseudomonas spp. have generally high intrinsic resistance compared with other Gram-negative bacteria. P. aeruginosa is often used in disinfectant testing because of its intrinsically high resistance to disinfectants [16] P. aeruginosa may adapt to high concentrations of QACs. The resistance mechanisms involved in the adaptation process is not fully understood [12].
Study of MICs for BKC demonstrated that P. aeruginosa strains were able to grow at a concentration of 500 µg/mL BKC while only two isolates (10% of total P. aeruginosa) were inhibited at a concentration of 300 µg/mL. The entire competitive flora was suppressed by <200 µg/mL BKC. Since two of the P. aeruginosa strains were sensitive to 300 µg/mL BKC, optimum BKC concentration was determined as 200 µg/mL to retain two P. aeruginosa (Table 2). These results showed that P. aeruginosa was more resistant to BKC than P. fluorescens and other Gram negative bacteria. Abraham et al. [17] informed that P. aeruginosa was resistant to some antimicrobial agents, as we observed.
3.3. Comparison of Selectivity of Two Media
Enumeration of P. aeruginosa and other Gram (-) bacteria were performed in CA and mCA and the results were shown in Table 3. 200 µg/mL BKC was added to CA to suppress the growth of competitive flora. After incubation, while the counts of P. aeruginosa strains remained constant with 0.68 log reduction, only two P. aeruginosa strains were determined sensitive to BKC and counts to be decreased by 5.4 log. In addition, the count of P. fluorescens strains were reduced 5.57 log. Counts of Acinetobacter baumannii, Enterobacter intermedium, Enterobacter agglomerans and Escherichia coli decreased 4.41, 4.77, 5.68, 7.10 log, respectively. These results indicated that 200 µg/mL BKC concentrations could inhibit of

Table 2. Minimum Inhibition Concentrations (MICs) for BKC.

Table 3. Enumeration of P. aeruginosa and competitive flora in CA and mCA (log cfu/mL)*.
competitive flora significantly, whereas 90% of all P. aeruginosa strains was not affected. When the two media were compared, the modified Cetrimide Agar was more selective than the standard CA for P. aeruginosa.
4. Conclusion
Selective media include Nutrient Agar supplemented with antibiotics, Cetrimide Agar, Pseudomonas Isolation Agar and growth media supplemented with C-390 (9- chloro-9-[4-(diethylamino)phenyl]-9,10-dihydro-10-phenylacridine hydrochloride) or with phenanthroline have been used for isolation of P. aeruginosa. But none of these supplements were a perfect selective agent. Some selective agents suppressed growth of Gram (-) bacteria as well as the growth of P. aeruginosa. To obviate this potential problem, in the current study BKC was used at a concentration of 200 µg/mL which suppressed the growth of other Gram-negative bacteria but does not have an effect on normal growth of P. aeruginosa. Our results showed that BKC modification to the Cetrimide Agar improved its selectivity by preventing the growth of other Gram-negative bacteria. This modified medium should be convenient medium to isolate of P. aeruginosa from raw milk.
5. Acknowledgements
The authors would like to thank Prof. Dr. A. Kadir Halkman to support this study.
NOTES