1. Introduction
Mycoplasmas are characterised by vastly reduced genomes and are among the smallest of the free-living organisms. Furthermore, mycoplasmas are diverse in terms of host environment, phenotypic features, as well as genomic characteristics [1] . They feature a reduced number of DNA repair proteins and exhibit high mutation rates, which contributes to the accelerated evolution observed within the genus [2] .
Most mycoplasmas are parasites, usually exhibiting strict host and tissue specificities. These organisms have evolved molecular mechanisms needed to deal with the host immune response including: mimicry of host antigens, survival within phagocytic and non-phagocytic cells and generation of phenotype plasticity. Mycoplasma hominis is an opportunistic human pathogen and resides as a commensal on the mucosal surfaces of the cervix or vagina of 21% to 53% of sexually mature, asymptomatic women; this is somewhat lower in the urethra of males [3] . However, M. hominis has also been associated with clinically diverse diseases including; urogenital diseases [4] [5] , postpartum fever [6] , pneumonia [7] , meningitis [8] [9] and septic arthritis [10] [11] . Although this organism has only been isolated from humans, it was found that in utero administration of M. hominis to pregnant macaque monkeys led to preterm labour and foetal lung injury [12] .
Mutation-based phenotypic and genetic variation is a strategy utilised by many pathogenic bacteria and protozoa to adapt to divergent host environments [13] [14] . These mutations often affect the surface structures of a pathogen and may therefore change functional aspects of the organism such as adherence, colonisation of the host, or immune evasion. High-frequency mutations, that are distinct from classical regulation of gene expression, are an adaptive tactic that can affect the expression or structure of selected gene products, creating and maintaining repertoires of functionally variant organisms within a population. This diversity may contribute in many ways to the survival, propagation, transmission and pathogenic properties of an infectious agent.
Mycoplasmas are heterogeneous organisms: many display antigenic variation and pronounced variation of surface proteins. This is thought to be an important way of evading the host immune response, particularly the humoral immune response, resulting in the chronic infection characteristic of many mycoplasma infections [15] . Previous studies have indicated that the surface antigenic profiles of M. hominis strains are highly heterogeneous expressing both size and phase variants of surface exposed membrane proteins [16] -[19] . Several M. hominis surface proteins have been characterised including P120, P135 and P50, however the molecular basis of variation in M. hominis has only been elucidated in some cases. The mechanisms involved in the diversification of mycoplasma surface proteins are highly complex and include: size variation caused by gain or loss of intragenic repetitive sequences; phase switching by deletion/insertion mutations or DNA inversion affecting promoter activity; and presence of multigene families or multiple copies of partial genes in the mycoplasmal chromosome [1] .
The presence of a variable adherence-associated surface protein of M. hominis was initially identified over 20 years ago as a potential adhesin of M. hominis by using specific monoclonal antibodies to inhibit mycoplasma adherence to cultured cells [17] [20] . This protein was first identified; as a 49 kDa surface protein in M. hominis strain PG21 [21] ; as an adherence-associated, multiple-banding membrane lipoprotein in strain 1620 [17] ; and as a 50 kDa adhesin in strain FBG, known as P50 [20] . While these groups used different nomenclature (Vaa versus P50), the surface lipoprotein in question is the same protein. We will use the original terminology, variable adherence-associated (Vaa) antigen, for the rest of this review as this lipoprotein often has varying molecular masses with conserved regions; therefore P50 is a less accurate descriptor. Variation in the composition and size of the Vaa proteins results from allelic variant forms of the single copy vaa gene in M. hominis [22] [23] . Despite discovery for more than two decades no comprehensive review of Vaa regulation and expression has been previously undertaken, the importance of Vaa in the pathogenesis and immune evasion of M. hominis infections is still not fully elucidated.
2. Phase Variation of the vaa Gene
Tandem repeats (TRs) are nucleotide sequences that are directly repeated in a head-to-tail manner. According to the conservation of the repeated sequences, TRs are classified as identical/perfect TRs or degenerated/imperfect TRs. Furthermore, TRs can be classified into three categories according to the size of the repeated unit: microsatellites, minisatellites and macrosatellites [24] [25] . The term “satellite DNA” originally refers to the very large arrays of tandemly repeated non-coding DNA that are characteristic of eukaryotic genomes however in the context of bacterial genomes the term is also used to include small and intragenic TRs [26] . In comparison to other bacterial species, several mycoplasma species, including M. genitalium, M. gallisepticum, and M. hyopneumoniae, contain long trinucleotide repeats in their genomes at a higher prevalence than is observed in other bacterial species. These repeat regions occur mainly in intragenic regions in the two former species and within coding regions of the latter [27] . In M. hyopneumoniae the trinucleotide repeats are located within hypothetical open reading frames or defined adhesins in which the gain or loss of these repeats results in variability of amino acid sequence. These changes in protein size and structure are speculated to influence protein-protein interactions and adhesion [27] .
There is abundant evidence that intragenic and intergenic TRs can promote phase variation. However, the underlying mechanisms are dependent on the nature of the TR. For example, if the TR unit size is not a multiple of three, rearrangements are able to induce frame-shift mutations causing ON/OFF phase variation of down-stream sequence (truncation) [26] . Phase variation refers to reversible molecular switches encoding ON/OFF gene expression resulting in variation in expression of one or more open-reading frames between individual cells of a clonal population. The frequency of phase variation is characteristic for the gene, the bacterial species, and the regulatory mechanism resulting in modulation of the switching frequency. Phase variation results in phenotypic variation within bacterial species [28] . Differences in the expression of Vaa between strains of M. hominis have been observed and Vaa has been shown to undergo high-frequency phase variation resulting in ON/OFF expression [17] [22] . Sequence differences have been observed between Vaa positive (Vaa+) and Vaa negative (Vaa−) variants derived from a single clonal lineage, with a single nucleotide deletion observed in a short tract of adenine residues (poly-A tract) located 166 nucleotides downstream of the ATG start codon. This tract contains eight alanine residues in cells expressing the full-length Vaa protein [23] . The deletion observed in Vaa− variants creates a frame-shift resulting in an in-frame UAG stop codon downstream of the poly-A tract.This mutation causes premature termination of translation and prevents Vaa expression [22] . Correction of this mutation has been observed in Vaa+ variants derived from a Vaa− clonal population, restoring the eight A residues in the polyA tract, and resulting in the expression of the full-length Vaa [22] . Thus phase variation of Vaa is controlled at translation, not as a consequence of transcriptional events due to promoter sequence divergence unlike transcriptional modulation of frame-shift mutations seen in other bacteria such as Necesseria gonorrhoeae and Ureaplasma sp. [22] [29] -[31] .
3. Size and Antigenic Variation of vaa
Size variation has been observed in Vaa using the monoclonal antibody H3 that was used to initially identify this protein, can inhibit the growth of M. hominis, and blocks attachment to host cells [17] . The size of Vaa observed in different isolates ranged from 28 kDa to 72 kDa and resulted from the gain or loss of intragenic repetitive sequences. Size variation of Vaa was initially examined in clonal lineages generated from M. hominis strain 1620, wherein three size variants of the Vaa antigen were identified and designated Vaa-2, Vaa-3, and Vaa-4. Sequence analysis showed that this vaa gene length variation corresponded to the number of 363 bp intragenic TR elements and the number of repeats was then used in the nomenclature of the different clonal lineages: Vaa-2 (two repeats), Vaa-3 (three repeats), and Vaa-4 (four repeats) [23] . These repeats form the basis of “modules” which provide a platform for further separation of Vaa types into categories (Figure 1). This 363 bp repeat became the prototype for what is now referred to as module III.
As shown in Figure 1, the first 240 amino acids of the FBG (P50) and 1620 Vaa proteins are highly homologous (96% amino acid identity), both of which contain a module III, however, divergence of the downstream sequence required the classification of further modules [32] . Sequence homology for each module type was initially restricted to 82% between strains [23] [32] . All vaa genes described thus far all start with highly homologous modules I and II, followed by either module II’ or II’’. All reported combinations of identified module types analysed in over 100 clinical isolates [32] [33] has resulted in six possible categories (Figure 1); however, the number of module III in category 4 have been found to vary between two and four repeats.
Module I contains 27 amino acids that encode the putative prolipoprotein signal peptide of the precursor protein [34] . This signal peptide is cleaved off in the mature Vaa, and the resulting N-terminal cysteine is lipid-
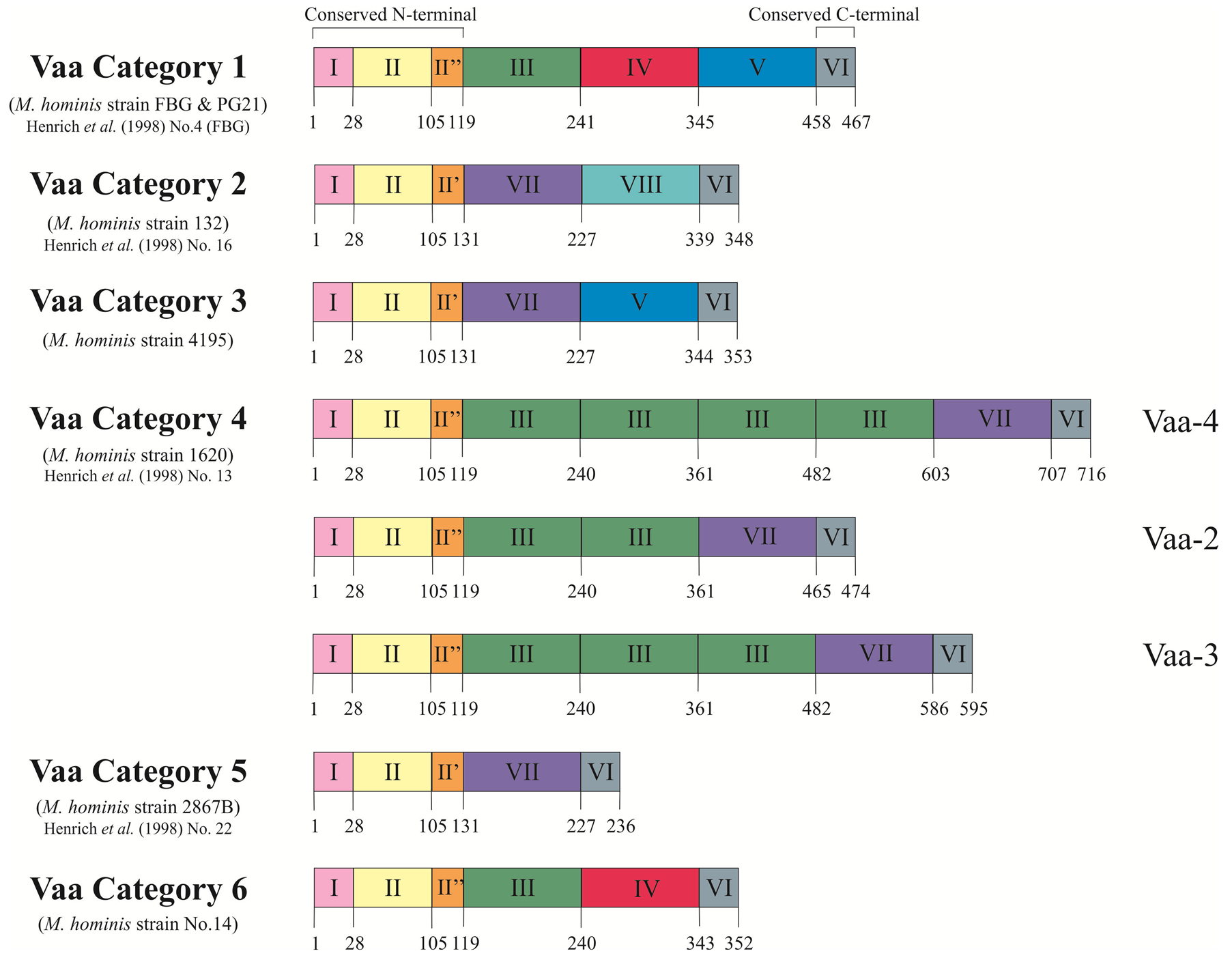
Figure 1. Schematic representation of the deduced amino acid sequences of the six vaa gene types. The proteins show a modular composition with homologous modules showing more than 82% amino acid identity. Modules I, II and I’/II” form the conserved N-terminal of the protein and Module VI represents the 10 amino acids conserved at the C-terminal. Modules III, IV, V, VII and VIII form the interchangeable cassettes. Prototype M. hominis strains for each Vaa category are stated along with corresponding strains from Henrich et al. (1998) [33] . Figure modified from Boesen et al. (1998) [32] .
modified, allowing the protein to be anchored to the bacterial membrane [35] . Module II encodes the conserved N-terminal end of the mature protein. Due to low sequence homology in the C-terminal end of this module, the amino acid region 105 - 118 has been split into two further modules, module II’ and module II’’. Module VI encodes the conserved 10 amino acids at C-terminal of all reported Vaa proteins.
Modules III, IV, V, VII and VIII form an interchangeable set of sequences that provide the size variation observed in Vaa variants. Inter-module homology (38% - 78%) suggests a common ancestral sequence [32] . A stable, repeated motif of four amino acids (SFKE) was observed in module II, a constant part of the gene. This motif was extended to ELESFKE in almost all of the interchangeable cassettes (identified by arrowed regions in Figure 2 and Figure 3). Three highly conserved tryptophan residues were also identified in distinct positions in a 16 amino acid region situated in the C-terminal part of the cassette sequence (identified by triangles in Figure 2 and Figure 3) [32] .
Using the previously proposed method of characterising Vaa type based on module composition as in Figure 1, we analysed 12 UK M. hominis strains that were collected between 1983 and 2012. The entire vaa gene was sequenced and the amino acid composition predicted from the open-reading frame (methods given in supplementary appendix). Eight of the UK strains belonged to category 1 and the remaining 4 were determined to belong to category 2 (alignments shown in Figure 2 and Figure 3). Aligning UK and prototype strains for category 1 Vaa showed a very high homology for modules I, II, and II”. The UK2012c strain had five unique polymorphisms through this region compared to the other strains, and it is interesting to note that this colonypurified strain originated from the same patient sample as UK2012b. This suggests that while Vaa type may be conserved within isolates from the same sample, micro-polymorphisms can exist within the same isolate. Of further note, nucleotide homology between these two strains for ten other essential genes showed 100% identity (data not shown), suggesting that Vaa may be more prone to mutation. In all strains analysed, the SFKE and ELESFKE motifs were conserved in module II and in the interchangeable cassettes, respectively. The conserved tryptophan residues can also be observed in the interchangeable cassettes. In the strains analysed, the modules that contain the highest levels of variation occur at the C-terminal end of the interchangeable cassette region, module V and module VII in Vaa category 1 and Vaa category 2, respectively. In fact, module V fails to maintain the 82% homology between isolates suggested by Boesen et al. (1998) to be required for inclusion in a particular module [32] . C-terminal modules appear to be hyper-variable in comparison to the other modules of the protein, indicating that there may be higher selective pressure to vary this region of the protein, suggesting that the C-terminus of Vaa may be more important to immune surveillance than more membrane-proximal modules. An-
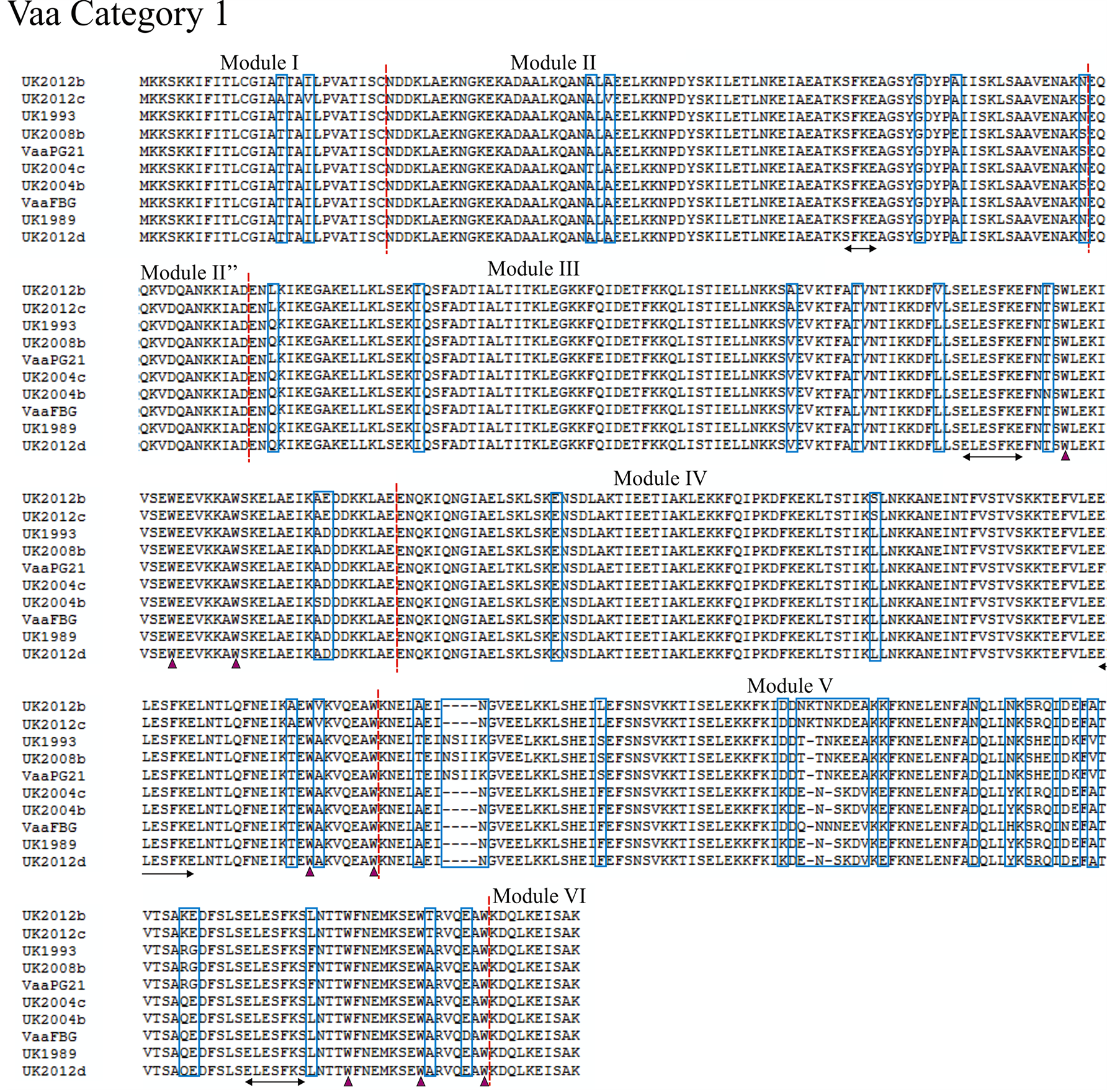
Figure 2. Alignment of the deduced amino acid sequence of Vaa category 1. Amino acid sequences of eight UK M. hominis strains and two prototype M. hominis strains (FBG and PG21) are shown. The modular composition of the protein is indicated and polymorphisms are highlighted by a blue box. The conserved SFKE and ELESFKE motifs (arrowed region) are observed in all proteins. Three tryptophan residues (triangle) are also conserved in the interchangeable cassette sequences.

Figure 3. Alignment of the deduced amino acid sequence of Vaa category 2. Amino acid sequences of four UK M. hominis strains and a prototype M. hominis strain (132) are shown. The modular composition of the protein is indicated and polymorphisms are highlighted by a blue box. The conserved SFKE and ELESFKE motifs (arrowed region) are observed in all proteins. Three tryptophan residues (triangle) are also conserved in the interchangeable cassette sequences.
tigenic variation is important to the expression of functionally conserved moieties within a clonal population that are antigenically distinct [28] .
The presence of these highly homologous interchangeable cassettes in the vaa gene suggests a mechanism of variation in which homologous recombination provides insertions or deletions of whole cassettes. This is most obvious in the Vaa-2, Vaa-3, and Vaa-4 variants isolated from a common ancestor (strain 1620). Homologous recombination can bring mutations arising in different genomes together and has a strong impact on pathogenic adaptation [36] . Homologous recombination was found in the penicillin-binding-proteins (PBPs) of Streptococcus pneumoniae, N. gonorrhoeae and N. meningitides and resulted in a mosaic gene structure [37] -[39] . Sequence blocks in the class A genes of resistant strains of S. pneumoniae confer decreased affinity to penicillin however these sequence blocks also contain mosaics of sequence similar to the sensitive strains. These blocks were thought to arise by interspecies horizontal genetic transfers followed by homologous recombination [40] . The class I outer membrane protein of N. meningitidis displays evidence of homologous recombination following intraspecies horizontal gene transfer resulting in the exchange of variable domains giving rise to antigenic variation of this protein [41] . The absence of sequences homologous to the vaa gene in other members of the mycoplasma family or other bacterial species suggests that intraspecies genetic transfer is responsible for the current array of Vaa categories.
4. Secondary and Tertiary Structure of Vaa
Sequence analysis and modelling of the Vaa protein indicates that Vaa belongs to the group of monomeric microbial surface-exposed coiled-coil proteins similar to Protein A of Staphylococci [35] [42] [43] . Vaa axial shape ratios indicate that the C-terminal region of the protein is elongated whereas the N-terminal region is globular. The secondary structure of Vaa examined by circular dichroism spectra and Jpred2 analysis indicated a primarily α helical structure with a predicted N-terminal region containing three α-helices interrupted by short breaks in helicity. Jpred2 analysis of the cassette region of the protein predicted two α-helices followed by two β-sheets and an α-helix. The secondary structure prediction of the C-terminal region of the protein implies the presence of two α-helices separated by a β-sheet [35] .
A hypothetical model, Figure 4, of the topology of Vaa shows a bacterial membrane lipid anchor that is typical of prokaryotic lipoproteins attached to the N-terminal cysteine residue of the mature Vaa with the conserved N-terminal in a triple-helix bundle, extending into an elongated helix. Two β sheets then form a loop region and a C-terminal helix folds back on the elongated helix. This model indicates that Vaa is composed of an N-terminal base domain in close proximity to the membrane and a C-terminal spike cassette domain projecting out from the surface of M. hominis [35] . The Vaa protein is characterised by its modular structure with different numbers
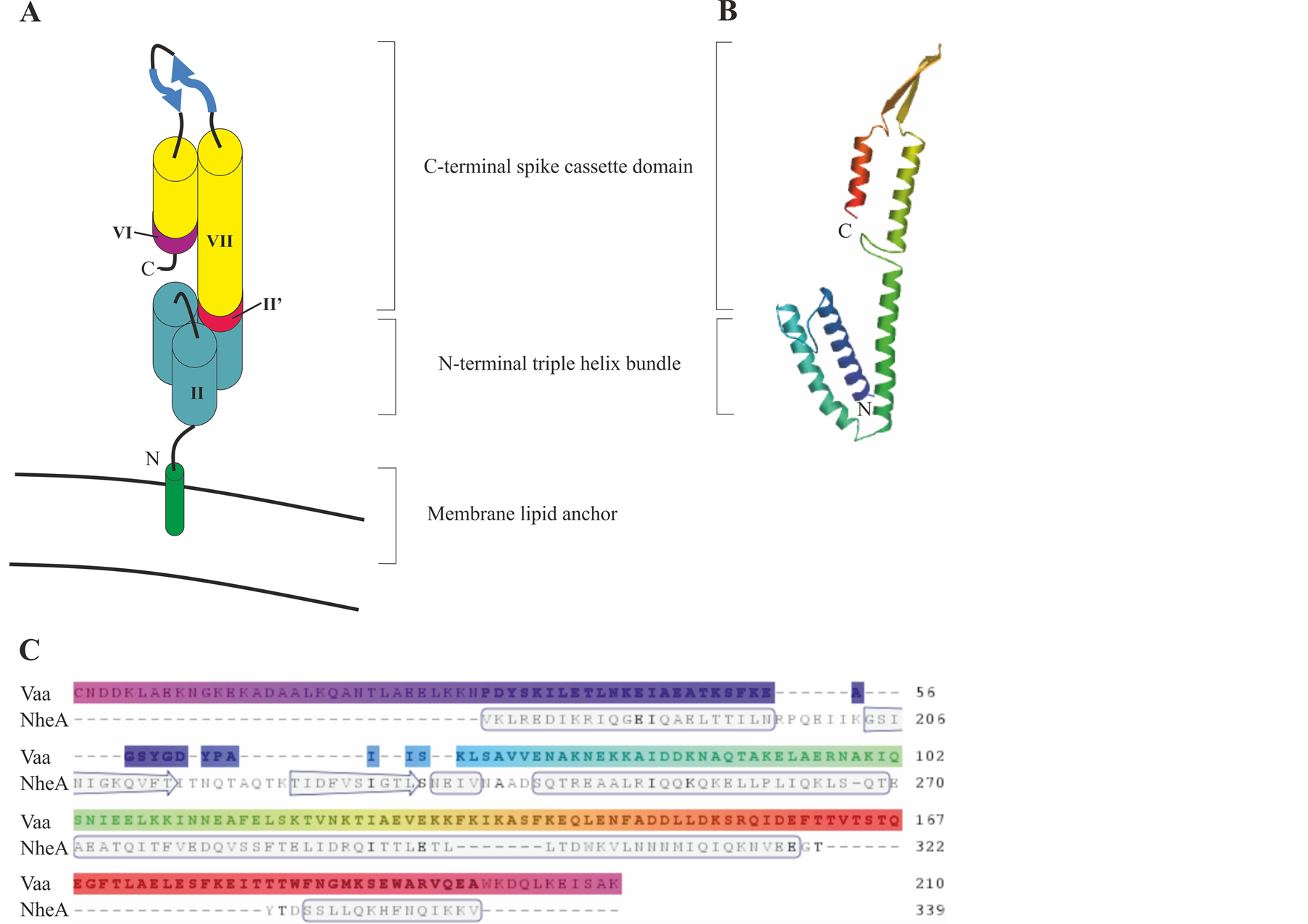
Figure 4. Model of the predicted protein structure for Vaa category 5. (A) Schematic representation of Vaa category 5 modified from Boesen et al. (2001) [35] . The modules are numbered as outlined in Figure 1. (B) Protein homology model of Vaa category 5 strain 2867B. Model was created using www.swissmodel.expasy.org and uses NheA protein as a template. (C) Alignment of Vaa category 5 strain 2867B amino acid sequence with the NheA protein template. Secondary structure is indicated in the template.
of interchangeable cassette sequences. It was originally proposed that the addition of a cassette could create a more elongated protein. However, analysis of a Vaa category 3 protein and a Vaa category 5 protein has revealed that the axial shape ratios of these two proteins, determined by circular dichroism, are almost identical, indicating that the interchangeable cassettes were arranged in parallel and not end-to-end [23] [35] .
5. Role of Vaa in Cyto-Adherence
Mycoplasmas have small genomes and limited biosynthetic capabilities, restricting them to a parasitic existence in association with eukaryotic cells of their host [3] [44] . The ability of mycoplasmas to adhere to host epithelial cells on mucosal surfaces, in the case of M. hominis the urogenital tract, is an essential stage to establish successful colonisation. Several mycoplasmas, such as M. pneumoniae and M. genitalium, have adhesin proteins associated with adhesion concentrated at specific tip structures [45] . In comparison, M. hominis, along with other mycoplasma species, lack this attachment organelle. Many surface antigens have been identified in M. hominis and some of these play a role in cyto-adherence as shown by monoclonal antibody inhibition assays [17] [20] .
Vaa has been shown to be involved in the adherence of M. hominis to host cells. Vaa was identified as a potential adhesion of M. hominis using monoclonal antibody inhibition, with the masking of Vaa (P50) showing prominent difference in the ability of M. hominis to adhere to HeLa cells [17] [20] . The role of Vaa as an adhesin of M. hominis was further investigated by determining which region of a category 1 Vaa protein was involved in adhesion to host cells [46] . Adhesion to glutaraldehyde-fixed HeLa cells with module III, modules III + IV, modules III + V (truncated proteins expressed in Escherichia coli) indicated the adherent property was distributed over the entire molecule, not localised to a specific region. However, adherence was increased when examining multiple modules [20] [46] .
This is further supported by the markedly reduced cyto-adherence of truncated Vaa proteins to cultured HeLa cells compared to the complete protein. Phase variation of the Vaa gene in a clonal lineage of M. hominis 1620 results in a mutation in the N-terminal of the gene and the production of a truncated form of the Vaa protein (Vaa−). This Vaa− variant showed a >70% reduction in adherence to HeLa cells compared to a variant expressing the full length protein [22] . Examination of the membrane protein profiles of both the Vaa− and Vaa+ variants revealed that the only detectable difference was the presence or absence of the Vaa protein indicating that the difference in adhesion was directly attributed to the expression of this protein [22] . Low residual adhesion of the Vaa− variant could be attributed to non-specific interactions between M. hominis and HeLa cells or from additional, unidentified adhesins [22] . However, adherence of a recombinant peptide containing the N-terminal region of category 1 Vaa has been demonstrated indicating that even the Vaa− variant may retain the ability to adhere through the N-terminal region of the peptide [46] .
6. Conclusion
The ability for M. hominis to adhere to host cells is a crucial step towards colonisation of a host. The Vaa antigen is a major adhesin of M. hominis and displays pronounced mutational variation in size as well as sequence and antigenic variation. To date the only mechanism described of altering Vaa expression relates to a truncation mechanism mediated by a poly-A (alanine-encoding) tract 161 bp down-stream of the ATG start codon, rather than a recombinase-mediated gene rearrangement such as described for phase variation in other mycoplasmas. Vaa truncation does, however, directly relate to the ability of M. hominis to adhere to host cells. Vaa displays a mosaic gene structure formed from interchangeable cassette sequences. Recombination of these cassette sequences results in different gene types thereby generating and maintaining functional diversity in M. hominis. Furthermore, methods of sequencing and characterising these distinct groups should be employed to determine if different Vaa modules are associated with isolation from different patient groups or sample sites. A method that begins the process of investigating differences between commensal populations and those that are associated with disease is overdue for this organism.
Supplementary Appendix
Materials and Methods
Isolates, Growth Media, and DNA
M. hominis isolates UK2012a, Uk2004a, UK2006, UK2008a, UK2012b, UK2012c, UK2004b, UK1993, UK2008b, UK2004c, UK1989 and UK2012d were submitted to Public Health England, U.K. for clinical diagnostic purposes. M. hominis isolates were grown in Mycoplasma Liquid Medium (Mycoplasma Experience, UK). Bacterial DNA from a 500 µl 48 hour culture was released by boiling lysis (95˚C for 10 minutes) following centrifugation at 13,000 x g for 10 minutes, removal of all MLM, and resuspended in 50 µl sterile water.
PCR Amplification
PCR was performed using the published primers of Zhang and Wise (1996) and the PCR conditions were performed as detailed by Boesen et al. (1998) [23] [32] . All the oligonucleotide primers were synthesised by invitrogenTM (UK) and the sequences of these primers are detailed in Table1 All PCR amplifications were performed in a DNA thermocycler (Techne Prime) in a volume of 50 µl containing: 1 x GoTaq® Flexi Buffer (Promega), 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 0.5 pM of each primer, 1.56 units of GoTaq® DNA Polymerase (Promega), and 2.5 µl DNA. The PCR products were analysed on 1.5% agarose gels with ethidium bromide visualisation. All PCR reactions were repeated twice.

Table 1. Primer sequences used for amplification of the vaa open-reading frame.
DNA Sequencing
PCR amplicons were purified using a Qiagen Mini Prep kit (Qiagen) as per manufacturer’s instructions and sequenced using the amplification primers, as performed by MWG Eurofins (Germany).
Sequence Analysis
NOTES
*Corresponding author.