1. Introduction
Food-borne diseases have increasingly become a health concern worldwide, with species of Salmonella, Campylobacter, Yersinia, Escherichia, Listeria and Clostridium being the most implicated [1 ]. Salmonella spp. have for long been reported as leading causes of food-borne infections [2]. In many settings, a further danger lies in the development of drug resistance in these organisms [2], primarily driven by non-prudent overuse of antiinfectives, presenting a serious threat to populations not only in resource-limited countries but globally due to increasing international travel and commercial transport where pathogens spread rapidly [3].
Because of the importance of Salmonella spp. as the cause of a food-borne disease, typing methods such as plasmid profiling, ribotyping and Pulsed-Field Gel Electrophoresis (PFGE) have been used to trace the outbreak to the contaminated source for public health intervention ADDIN EN.CITE ADDIN EN.CITE.DATA [1 ]. Plasmid analysis has recently been used to investigate an outbreak of multiresistant (R-type ACSSuSpT) Salmonella Typhimurium DT 104 in England and Wales [4] and field Salmonella isolates from poultry, humans, cows and poultry house environments in Iran [5]. In Uganda, Salmonella organisms occurred in stools of 8.1% of patients with acute diarrhea in Kampala district, with 69.2% of these isolates exhibiting multiple antibiotic resistance [6]. However, that study did not further characterize the isolates. The current study, therefore, aimed at determining the antimicrobial resistance patterns and plasmid profiles of Salmonella isolates from humans and those from foods of animal origin from different parts of Uganda so as to understand the extent of drug resistance, genetic diversity and possibility of cross species transmission of the organisms.
2. Materials and Methods
2.1. Study Isolates
A total of 58 human-derived Salmonella isolates (archived between July, 2007 and July 2009) were collected from Health Centre Laboratories across the country (16, 17, 12 and 13 isolates from Mbale, Lacor, Mulago and Entebbe hospitals respectively). Isolates were reidentified and additional information about socio-demographic characteristics of the patients retrieved from the Laboratory Information Systems. Additionally, 34 isolates previously isolated from foods obtained from common food markets around the country were collected from the Central Diagnostic Veterinary Laboratory, College of Veterinary Medicine and Biosecurity, Makerere University. Isolates from cattle, chicken, pigs, eggs and associated food products were analyzed because these animals/products are possible carriers of Salmonella organisms.
2.2. Identification and Antimicrobial Susceptibility Testing Salmonella identification was carried out according to a protocol from the Health Protection Agency (UK), 2007, in which microscopic appearance, growth on primary isolation media, colonial appearance, and biochemical reactions were used. The biochemical tests included urease, and the oxidase tests. The disc diffusion method (Kirby Bauer) was done in accordance to the National Committee for Clinical Laboratory Standards (NC-CLS) .The direct colony suspension method was used, and an equivalent of 0.5 McFarland concentration of innoculum was prepared from an 18 - 24 hour growth of pure colonies of Salmonella. Incubation was then done at 35˚C for 18 - 24 hours. Isolates were then regarded as susceptible, resistant or of intermediate resistance according to the zone of inhibition around the disc. Ampicillin (10 μg), chloramphenicol (30 μg), nalidixic acid (30 μg), tetracycline (30 μg), and ciprofloxacin (5 μg) were tested against according to the global Salmonella surveillance protocols of the World Health Organization [7] and also in accordance to the treatment guidelines of the Ministry of Health in Uganda [8].
2.3. Plasmid Profiling
Plasmid extraction was done using a QIAprep Spin miniprep kit (Qiagen, GmbH, Germany). A protocol adopted from the global Salmonella surveillance protocol of the World Health Organization was used [7]. Plasmids were separated by agarose gel electrophoresis with a molecular weight marker of a mixture of λ DNA/HindIII and φχ 174 DNA/HaeIII.
2.4. Data Analysis
Plasmid profile patterns were scanned and data entered into the BioNumerics software version 5.0 (Applied Maths NV, St. Matens Latem, Belgium) and analyszed as fingerprint types. The pair wise distances between patterns were computed using unweighted pair-group method using arithmetic averages (UPGMA) and the Jaccard index at a tolerance level of 2.0%. A cluster was considered to be at least two isolates with the same fingerprint pattern. Chi squares were computed in STATA version 11 and a P value of <0.05 was considered evidence of significant difference.
2.5. Ethical Considerations
The study used archived isolates at various reference laboratories in Uganda. No patient identifiers accompanied isolates. Permission to use the isolates was obtained from the Institutional Review Board of the School of Medicine, Makerere University, and the Uganda National Council for Science and Technology.
3. Results
3.1. Sources of Salmonella Isolates
A total of 92 isolates were analyzed. Of these, 58 were human-derived while 34 were from foods of animal origin. Most of the animal derived isolates (26/34) were from meat and meat products, 6/34 were from poultry while only 2/34 were from milk and milk products.
3.2. Antimicrobial Drug Resistance Patterns
All the 92 isolates were tested for antibiotic susceptibility. Among the human-derived isolates, 57/58 (92%, 95% CI, 91.8% - 99.9%) were susceptible to Ciprofloxacin while 32/34 (94.1%, 95% CI, 81.9% - 91%) of animal-derived isolates were susceptible to Ciprofloxacin. On the other hand, 48/58 (82.3%, 95% CI, 71.4% - 91%) human derived isolates were resistant to Trimethoprim Sulfamethoxazole compared to 29/34 (85.3%, 95% CI, 70.4% - 94.4%) of the animal derived isolates. Fifty four percent
(n = 50) of the all the isolates were resistant to at least three antibiotics whereas only 2.2% (n = 2) of the isolates were susceptible to all the seven antibiotics tested. Forty five percent of the Salmonella isolates were susceptible to all three drugs commonly used in treatment of salmonellosis (chloramphenicol, nalidixic acid and ciprofloxacin) while only 1% of the isolates were resistant to the three drugs. Detailed susceptibility results for both animal and human-derived isloates are shown in Figure 1.
The risk of ampicillin resistance was almost three times more likely to occur in animal-derived as compared to human-derived isolates (Odds Ratio = 2.705, 95% CI, 1.3 - 5.8); that to nalidixic acid was also three times more likely to occur in animal-derived as compared to humanderived isolates (Odds Ratio = 2.895, 95% CI, 1.17 - 7.2), while that to Ciprofloxacin in animal-derived isolates was not significantly different from that in human-derived isolates [P = 0.527 (CI, 0.19 - 24.6)]. Detailed susceptibility results are indicated in Table 1. The high risk of resistance to nalidixic acid (one of the drugs commonly used in treatment of salmonellosis) in animal-derived compared to human-derived isolates poses a dilemma in the treatment of salmonellosis in this setting.
3.3. Plasmid profiles of isolates
Twenty two of the isolates did not have plasmids. The rest of the isolates had at least one plasmid equal to or smaller than 24 kb. In the 67 isolates analyzed, strains possessed at least one to five plasmids which resulted in 35 different plasmid profiles. There was formation of eight clusters comprising of 68.7% (46/67) of the isolates. In five of the eight clusters formed, there were both animal and human isolates. The biggest cluster of 12 isolates contained 10 human-derived and two animal-derived isolates, while another cluster of six contained four human-derived and two animal-derived isolates (Figure 2), suggesting cross-species transmission.

Figure 1. Level of antibiotic resistance to the seven drugs tested. Amp = Ampicillin, Tet = Tetracycline, Chlor = Chloramphenicol, Nal = Nalidixic acid, Sxt = Trimethoprim Sulfamethoxazole, Cip = Ciproflaxacin, Cro = Ceftriaxone.

Figure 2. Plasmid profiles and clustering of the isolates with at least one plasmid (n = 67). ID = the study number of the isolate, Origin = source of isolate and Susceptibility = resistance or sensitivity to the drugs tested, in the following order: ampicillin, chloramphenicol, nalidixic acid, ceftriaxone, trimethoprim sulfamethoxazole, tetracycline and ciproflaxacin.
4. Discussion
Salmonella organisms are among the most common causes of human bacterial gastroenteritis worldwide, and food animals are important reservoirs of these bacteria [9]. In recent years, an increase in the occurrence of antimicrobial drug—resistant Salmonella species has been observed in several countries. Specifically, it is endemic to Southeast Asia, the Indian subcontinent, and South America and is a growing problem in Africa [10-12 ]. Developed countries are thus rarely challenged with salmonellosis, and cases are usually due to travelers return
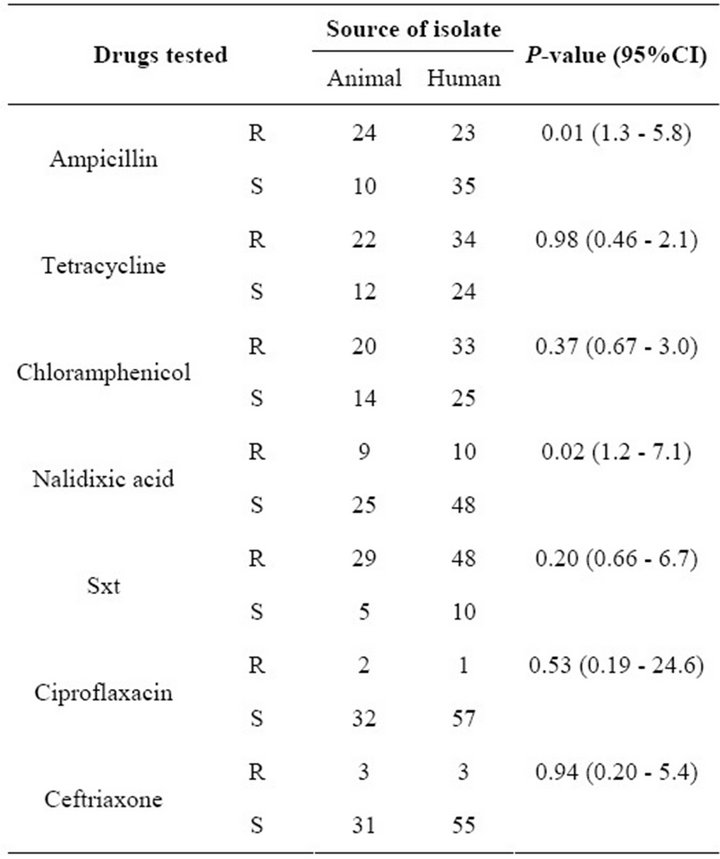
Table 1. Drug susceptibility of isolates from man and foods of animal origin. R = resistant, S = sensitive, Sxt = Trimethoprim Sulfamethoxazole.
ing from areas of high endemicity [9,13 ].
The current study compared the resistance pattern and plasmid profiles of Salmonella isolates from humans and foods of animal origin. We have found that 76% of meat and meat products in our sample were contaminated with Salmonella spp. This is in agreement with a study in peri-urban Kampala in 1998 found which reported that meat and meat products were the most common source of infection with Salmonella spp. in the city [6]. The most prevalent antibiotic resistance pattern to the commonly used drugs in the treatment of salmonellosis in this setting (Chloramphenicol, Nalidixic acid and Ciprofloxacin) was resistant, sensitive, and sensitive respectively, making Chloramphenicol highly ineffective. Only 45% percent of the isolates were susceptible to all the three drugs above, while 55% of the isolates were resistant to at least two of the three drugs. The implication of this finding is that current treatment options may have to be reviewed in light of this new pattern of resistance. Multi-resistance to the antibiotics tested was more pronounced in those isolates which had multiple and heavy plasmids, and we thus hypothesize that resistance in these isolates may have been plasmid mediated. All the drugs that are used in treatment of salmonellosis in man are also commonly used in the treatment of various conditions in animals, including use in processed animal feeds. With a common animal-human interface, antibiotic resistance may easily be transferred from animals to man. This may explain the marked increase of resistance to drugs like chloramphenicol and tetracycline in isolates of human origin observed in this study. In our setting, there is indiscriminate use of antibiotics in animals, with many farmers using very high doses of drugs to treat their animals, without veterinary guidance. For example salmonellosis in chicken in this setting is treated using 20% tetracycline, creating a big antibiotic pressure on these organisms, which are later transferred to man through consumption of poultry products. A study in health centers in neighboring Kenya [14] reported that 78.8% of Salmonella isolates of human origin were resistant to Chloramphenicol , which is close to what we observed here (Table 1), but their study found no resistance to Ciproflaxacin, which we have detected in both human and animal isolates, suggesting an ominous situation in our setting. revious studies have shown that the number of plasmids and related profiles in Salmonella enteritidis varied, with some studies reporting few patterns [5,15 ]. Our study, on the other hand, revealed 35 different plasmid profiles, with eight clusters ranging from two to 12 isolates each. The multiplicity of profiles may be due to presence of several serotypes, but we could not establish that in this study. Among the eight clusters, four had both human and animal-derived isolates, suggesting cross-species transmission between animals and man. A previous study in the Netherlands isolated S. typhimurium DT104A variant from a diseased pig, calf, and child on a single farm. All three strains were typed by phenotypic and genotypic methods and were identical, suggesting transmission across species [16]. Similar studies have been done in East Africa [6,14 ] but our study is the first, regionally, to compare plasmid profiles and resistance patterns of isolates from animal-derived foods and isolates of human origin, with results highlighting the extent of the public health dilemma caused by this pathogen. In the current study, clustered isolates also showed a common resistance pattern to at least three out of the seven drugs tested. While food handlers play a significant role in the transfer of Salmonella spp. from one person to another, it is also possible that in our setting, the close relationship between livestock and humans, in combination with poor animal waste disposal, may lead to washing of animal excreta into springs and other water bodies which thus become the source for infection to man.
5. Conclusion
The Study showed that meat and meat products are the commonest source of Salmonella infection in this setting. There was clustering of strain of human and animal origin, suggesting cross-species transmission of plasmids, and possibly drug resistance. The findings further provide evidence to agencies and legislators involved in making policy decisions about the use of antimicrobials for the need of guidelines for the prudent use of antibiotics in food animals in Uganda.
NOTES