Electrospun Nanofiber Membranes Containing Molecularly Imprinted Polymer (MIP) for Rhodamine B (RhB) ()
1. Introduction
Molecularly imprinted polymers are obtained by functional monomers and cross-linking monomers around a template molecule, leading to a highly three-dimensional cross-linked network polymer [1,2]. When polymerization by covalent, non-covalent or semi-covalent approaches has taken place and template molecule is removed. MIPs with binding sites with shape, size and functionalities complementary to the target analytes are established. MIPs have an unusual recognition of their corresponding target molecules [3,4]. Because of the selectivity, easy preparation and economic, MIPs are ideal materials and the recent concerns! The MIPs are stable, robust and resistant to a wide range of pH, solvents, thermal stability, and have good specific adsorption properties and selectivity ability. It has been widely used for selective enrichment and separation in environmental applications [5-7], food applications [8], chemical sensors [9-11], optical sensors [12], protein selections [13,14], and pharmaceutical analysis [15,16]. However there are several limitations and challenges with the rapid development of MIPs. For example, 1) MIPs are usually very small particles and hard to accumulate during the experiment; 2) the process column needs to be filled and large consumption of solvents; 3) some active sites were also embedded into the particles in the traditional methods. So the methods and strategies of easy preparing MIPs with much larger surface areas are important and urgent.
The ability to create materials with well-controlled nanofibers is of intense interest for a variety of applications. In recent years, electrospinning technology has gained widespread attention since it is known to be an effective fabrication tool for preparing various polymeric nanofibers and nanostructured materials with a high aspect ratio and high surface area [17-20] (Scheme 1). So electrospun nanofibers are good candidates for the adsorption and filtration because of their high permeability, large surface area and small pore size. In addition, the electrospun nanofibers are easy to assemble, modify and process into different applications, such as biomedicine [21], tissue engineering [22] and armored fabric.
Furthermore, the introduction of MIP on the surface of

Scheme 1. Schematic diagram of a laboratory basic setup for an electrospinning experiment with a horizontal arrangement of the electrodes.
an electrospun nanofiber membrane enables its utilization as an adsorbent in separation and accumulation for complex sample treatment. Keiichi Yoshimatsu and Lei Ye found that molecularly imprinted nanoparticles were encapsulated into polymer nanofibers with electrospinning method [23]. While Ioannis S. Chronakis and Lei Ye prepared encapsulated and selective recognition of molecularly imprinted theophylline and estradiol nanoparticles within electrospun polymer nanofibers [24]. Silvia Piperno described the polymer nanofibers with entrapped molecularly imprinted polymer (MIP) nanoparticles and studied their possible use in a fluorescence-based biosensor application [25]. Compared to encapsulation for nanoparticles, there are some advantages for molecularly imprinted polymers to dissolve into uniform and homogeneous electrospinning solutions. So Ioannis S. Chronakis and Lei Ye prepared smooth electrospun poly-(ethylene terephthalate) polymer which contained molecular recognition sites for 2,4-dichlorophenoxyacetic acid [26]. The electrospun nanofibers provided a porous matrix with very high surface area-to-volume ratio, which is desirable for the selective rebinding of the target molecule. They demonstrated for the first time that electrospun template directed molecular imprinting is a viable method for creating molecularly imprinted nanofibers.
Rhodamine B (RhB) is a kind of peach red synthetic dye. It is widely used in industry, such as textile, plastics, food, dyeing, paper making and printing. As a consequence of its solubility, high chemical and biological stability, RhB has been affected to environment [27,28]. It is well known, RhB also is harmful to skin, eyes and brain. So nowadays there are many ways to remove RhB, such as condensation, sink to the bottom, adsorption [29, 30], reversed-phase permeability, ion exchange and chemical oxidation [31]. But there is no mature and effective method to separate and accumulate RhB in some complex samples, such as food, environmental water samples. So we choose RhB as our research objective to develop some economical and effective separation ways!
Therefore, we use the precipitation polymerization to make RhB molecularly imprinted polymer particles. Then these MIMs through electrospinning method were made. The MIMs were used for separation and accumulation of RhB. The results showed that the imprinted polymer exhibited higher affinity compared to molecularly imprinted particles. Meanwhile it is necessary to identify several optimal conditions with respect to maximum separation performances.
2. Experiment Section
2.1. Materials and Instrumentations
Acrylamide, ethylene glycol dimethacrylatea (EGDMA), azobisisobutyronitrile (AIBN), acetonitrile and polyethylene terephthalate (PET) were purchased from J&K Scientific Ltd. Rhodamine B (RhB) was obtained from Sinopharm Chemical Reagent Co. All solvents were obtained from Sinopharm Chemical Reagent Co. and used as received. All aqueous solutions were prepared using purified water with a resistance of 18.2 MΩ·cm.
Field emission scanning electron microscopy (FESEM) was obtained by Hitachi S-4800 (S-4800, Hitachi Ltd., Japan). The HPLC (Diane 3000) and UV/Vis (Shimadzu 2550) were used for analyses. High voltage direct current generator (JG50-1) was bought from Shanghai Shenfa Ltd.
2.2. Synthesis of the Electrospinning MIP Membranes
2.2.1. Preparation of Molecularly Imprinted Polymers (MIPs) Microspheres
A mixture containing 0.5 mmol RhB, 3 mmol Acrylamide, 15 mmol EGDMA and 100 mg AIBN were placed in a glass vial. After ultrasonic mixing for 5 min, the mixtures were purged with nitrogen for 15 min. The polymerization proceeded at 60˚C for 24 h. Then the template molecules were washed away by mixed solvent (acetic acid: methanol = 1:9 (volume)), until no RhB could be detected in the washing solutions using UV/Vis spectroscopy. After vacuum drying, MIP microspheres were obtained.
2.2.2. Preparation of Molecularly Imprinted Membranes (MIMs) via Electrospinning
Polyethylene terephthalate (PET) solution was prepared by adding PET (1.0 g) into trifluoroacetic acid (TFA) (4.0 mL) with continuous agitation in close vials for 3 h at room temperature. Then the solution of molecularly imprinted polymers in the dichloromethane was blended into PET solution. After agitation and ultrasonication for 2 h, the resultant homogeneous solutions were fed into a plastic syringe driven by a syringe pump (KDS-200, Focus Co., Ltd., USA). The positive electrode of a high voltage power supply was clamped to the metal needle (0.8 mm) tip of the syringe. The cylindrical collector covered by aluminum foil was used for collector. The applied voltage was 18 KV, and the tip-to-collector distance was 15 cm. The ambient temperature and relative humidity were maintained at 25˚C and 45%, respectively. The prepared fibrous membranes were dried in vacuum at room temperature to remove the trace solvent. NIMs were prepared by the same process without RhB for template. The morphology and size of polymer microspheres and electrospinning membranes were obtained by scanning electron microscopy.
2.3. Analytical Conditions
2.3.1. Optimization of the Amount of MIP Membranes
Molecularly imprinted polymer membranes of 10.0 mg, 20.0 mg, 30.0 mg, 40.0 mg, 50.0 mg and 60.0 mg were added into RhB aqueous solution (40 μmol/L, 10 mL). Then the sample was detected and analyzed after slight shock in continuous rotary oscillator for 2 h. The solution of 10 μL was removed for the analysis with HPLC each time.
2.3.2. Effects of pH
BR (Britton-Robinson) buffer solution was prepared with different pH values to research RhB adsorption situation.
2.3.3. Optimization of Adsorption Time
To investigate the adsorption time, molecular imprinted polymer membranes were added into Rhodamine B aqueous solutions on the rotary oscillator to slightly shock. From the beginning time, a sample was taken to detect and analysed with HPLC every 10 min. NIMs were also investigated in the same method to determine the adsorption time compared to the corresponding MIMs.
2.3.4. Static Adsorption
After optimization of the amount of membrane, 10.0 mg MIMs were selected as adsorbents in the process of the static adsorption experiments. MIMs were added into Rhodamine B aqueous solution with different concentrations (1 - 40 μmol/L). After slightly shocked at room temperature in the rotary oscillator for 2 h, the samples were analysed by HPLC. NIMs were also studied in the same method.
2.3.5. Chromatographic Conditions
The concentrations of Rhodamine B solutions were assayed on UltiMate 3000 high performance liquid chromatography (HPLC; Diane, USA). The HPLC system equipped with a G1314B UV/Vis detector and G1328B manual-sampler. The stationary phase consisted of a column (5 μm, 4.6 mm × 250 mm) packed with extend-C18. The detection wavelength was 550 nm for Rhodamine B. The mobile phase consisted of watermethanol (25:75, v/v) for adsorption, water-methanol (28:72, v/v) for pH effect, and water-methanol (65:35, v/v) for river water sample. The column was eluted at a flow rate of 1.0 mL/min at 35˚C.
2.3.6. Recovery Experiments
Samples with high, medium and low concentrations were prepared by the stock solution (6 μmol/L of RhB) and RhB simulated water samples. 70%, 50%, 30% of the amount of reference substance stock solutions (6 μmol/L) were added into water samples (20 mL) to make recovery experiments separately for the imprinted membrane and blank membrane.
MIMs (10.0 mg) were collected to place into conical flask. Then add to the high, medium and low concentration samples. After slight shock for 2 h at room temperature in a rotary shock device, liquid chromatography was used to determine the equilibrium adsorption concentration of the solution. According to the concentration of the solution before and after adsorption, the changes in the molecularly imprinted polymer membranes for the adsorption of RhB target molecule were calculate. The same method was used to determine the adsorbed NIMs.
Unknown concentrations of RhB simulated water sample was made in the range of standard curve. Through pre-treatment with the imprinted membrane, the concentration of samples was detected by liquid chromatography. At the same time NIMs were used to determine and then compared with the imprinted membrane.
3. Results and Discussion
3.1. Preparation of Molecularly Imprinted Microspheres
Precipitation polymerization was used to prepare molecularly imprinted microspheres (Scheme 2). In the reaction system, surface of prepared polymer microspheres was clean because of without the stabilizing agents or surfactants. SEM observation found that the polymer microspheres were spherical, and were arranged in an orderly three-dimensional shape. There is only a little bonding between the particles (Figure 1). The diameters of molecularly imprinted microspheres (a) and nonmolecularly imprinted microspheres (b) were about 1 mm.
3.2. Preparation of Molecularly Imprinted Membranes via Electrospinning
In our work, PET solution prepared by adding PET into trifluoroacetic acid (TFA) with continuous agitation was used for electrospinning. PET was selected as the sup-

Scheme 2. Schematic diagram of the molecular imprinting process for RhB.
 (a) (b)
(a) (b)
Figure 1. SEM images of molecularly imprinted microspheres (a) and non-molecularly imprinted microspheres (b).
porting fiber matrix because it could be easily electrospun to ultrafine nanofibers [26]. The solution of molecularly imprinted polymer (MIP) in the dichloromethane was blended into PET solution. SEM observation found that the MIP wire and blank NIP wire show smooth surfaces and good morphology. The long, electrospun nanofibers were randomly distributed with very uniform and dense structures. The fibers exhibited good morphology with an average fiber diameter in the range of 100 - 200 nm (Figure 2). The addition of RhB template molecule to the starting polymer solution caused no noticeable change in morphology in the nanofibers. It was based on the fact that the electrospinning started from a homogeneous polymer solution containing PET, and RhB template molecularly imprinted microspheres.
3.3. Adsorption Experiment
In order to future applications, we were interested in studying RhB binding in an aqueous environment at a low concentration level. Adsorption experiments were operated by MIM or NIM at different concentrations of
 (a) (b)
(a) (b)
Figure 2. SEM images of electrospinning MIMs (a) and NIMs (b).
target molecules (or template molecules), respectively.
The adsorption capacity Q is calculated by the following formula:
 (1)
(1)
where Q is the static equilibrium adsorption capacity (mg/g); C0 is the initial substrate concentration (mg/L); C is the substrate concentration (mg/L) at equilibrium; V is the volume of substrate solution (L); m is the amount of added polymer film (g).
3.3.1. Optimization of the Amount of MIMs
Optimization of the amount of film is to examine the optimum amount of film to achieve the province of the material and manpower. From Figure 3, we can see in the same conditions, the adsorption capacities were reduced with the increase of the membrane quality. The amount of 10 mg MIMs was selected in the following experiments based on the economic point.
3.3.2. Effects of pH
RhB solutions with different pH (3 - 10) were prepared to research RhB adsorption situation. Then 10 mg MIMs was added into the solution. The samples were collected after slightly shake for 2 h. Then UltiMate 3000 series of HPLC (UV detector) was used for detection of RhB. In Figure 4, the results showed that the adsorption of RhB is affected by pH.
Carboxyl was damage by esterification under acidic conditions (pH < 7). Conversely the hydrogen bond of the amino was lost under alkaline conditions (pH > 7). So the recognition sites on the polymer were destroyed. And then the adsorption properties of the MIMs were affected. So the neutral condition (pH = 7) is the best conditions in

Figure 3. The dosage optimization of MIMs for adsorbance of RhB.

Figure 4. The dependence between adsorbance and pH for MIMs in absence of RhB.
the later studies.
3.3.3. Optimization of Adsorption Time
Optimization of adsorption time was to determine the best time in adsorption process. From Figure 5, the adsorption of MIMs was superior to the NIMs and had higher recognition ability than NIMs. At the same time, MIMs and NIMs has a desorption phenomenon after the slightly shock for 80 min. So the samples were extracted after slightly shock for 80 min for analysis in the followup work.
3.3.4. Static Adsorption
From Figure 6, we can see with the concentration increase of Rhodamine B, the adsorption of RhB was increased by the same quality of Rhodamine B MIMs. Meanwhile the adsorption of RhB of MIM was obviously superior to the NIMs. So this phenomenon can illustrate that the MIMs contains a lot of specific binding sites.
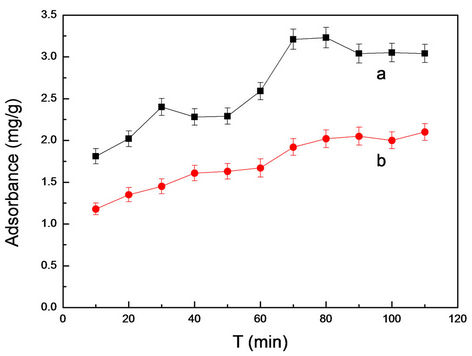
Figure 5. The dependence between adsorbance and time for MIMs (a) and NIMs (b) in absence of RhB.

Figure 6. The dependence between adsorbance and concentrations of RhB for MIMs (a) and NIMs (b).
3.3.5. Regeneration of MIMs
The stability and reproducibility of MIMs were investigated. As shown in Figure 7, the adsorption of MIMs for RhB was not significantly reduced after eluting several times (n > 5) with pure methanol. Therefore, the results show that MIMs has good regenerative adsorption and availability.
3.3.6. Elution and Elution Volume
Different eluents have different eluotropic capacities for target molecules. This paper studied the eluotropic capacities of methanol, acetonitrile and water. The eluotropic capacities between methanol and acetonitrile were equivalent. Because of the toxicity of acetonitrile, methanol is better than acetonitrile. From the other side of eluotropic time, the eluotropic time of methanol (0.5 h) is shorter than water (2 h). Therefore, from the above considerations, the chromatographic pure methanol acted as eluent in the experiment.
3.4. Analytical Performance
3.4.1. Standard Curve and Detection Limit
A series of low concentration of the RhB standard solution were prepared by using solution of RhB, which analyzed by HPLC method with the optimized chromatographic conditions. And the standard curve could be expressed by the peak area (A) on the concentration (μg/mL). The results indicated that the linearity was investigated at the range of 0.01 - 20 μmol·L−1 with squared coefficients of correlation R = 0.9996. And the linear equation was A = 1.91*C – 0.2. Additionally, the detection limit (LOD; 3 σ/K, n = 11) for RhB was found to be 2 nmol·L−1.
3.4.2. Evaluation of Method
The high, medium and low concentrations of three dif-

Figure 7. The adsorbance changes between different applications for the same MIMs for RhB.
ferent standard RhB solutions were added in the river water samples. MIMs or NIMs was used to enrichment the RhB in river water samples for 1 h with the optimal experimental conditions. Then the MIM or NIM was washed with methanol for three times. And the spiked recoveries were determined by HPLC. From Table 1, the recovery of MIMs for RhB was 97.80% - 117.2% and the relative standard deviation (N = 3) was 1.4% - 8.7%. HPLC results showed that there is no RhB in river water sample. And the recovery of MIMs is obviously higher than NIMs. So MIM has good selective adsorption for RhB than NIMs.
4. Conclusion
In this paper molecularly imprinted polymer particles of Rhodamine B was synthesized. Then molecularly imprinted membranes (MIMs) were made by electrospinning technology. MIMs were applied to enrichment and separation of RhB. All results show that MIMs have good selective adsorption than NIMs and achieve the desired effect. The above results demonstrate that electrospun molecular imprinting is a viable method for creating robust molecularly imprinted nanofibers that can selectively the RhB target molecule. The imprinted nanofibers also had a well-defined morphology and physical stability. Considering nanofiber processing, the present electrospinning-imprinting method is promising for its simple operation and high efficiency. Meanwhile, we have prepared the magnetic molecularly imprinted polymers using atrazine as a template on surface modification of the Fe3O4@SiO2 nanoparticles [32]. So we are designing the molecularly imprinted membrane based on the electrospinning technology and magnetic materials.
5. Acknowledgements
This work was financially supported by the Natural Sci-
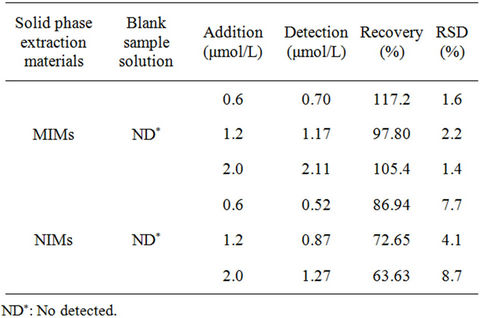
Table 1. The recovery of standard addition and relative standard deviation (RSD) with RhB in river water sample.
ence Foundation of China (No. 21177049, No. 5110- 3063), the Zhejiang Provincial Natural Science Foundation (No. Y4110545), the Program for Science and Technology of Zhejiang (No. 2011C22096, No. 2011- C37033), the Zhejiang Science and Technology Innovation Project for university student (2011R417020) and the Program for Science and Technology of Jiaxing (No. 2010AY1081, No. 2011AY1028, No. 2011AY1007).
NOTES