Phenotypic Detection and Susceptibility Pattern for the Detection of Extended Spectrum β-Lactamase-Producing Klebsiella pneumonia Isolates in Nairobi, Kenya* ()
1. Introduction
The genus Klebsiella belongs to the tribe Klebsiellae, a member of the family Enterobacteriaceae. The organisms are named after Edwin Klebs, a 19th century German microbiologist. Klebsiellae are non-motile, rod-shaped, gram-negative bacteria with a prominent polysaccharide capsule. Klebsiellae are ubiquitous in nature. In humans, they may colonize the skin, pharynx, or gastrointestinal tract. They may also colonize sterile wounds and urine. Klebsiellae may be regarded as normal flora in many parts of the colon and intestinal tract and in the biliary tract. Klebsiella species cause various infections including pneumonia, septicaemia, bacteraemia, meningitis, osteomyelitis, wound infections, urinary tract infections, childhood gastroenteritis and other conditions.
The discovery of penicilins was later accompanied by the discovery of penicillin destroying enzymes initially referred to as penicillinases and subsequently as β-lactamases. Enzymes are the major targets whose alteration results in resistance. Many antibiotics are structural analogues of natural metabolites and therefore inhibit enzymes that recognise the antibiotics or their metabolites as substrates [1]. Beta-lactamases are serine proteases which catalyse hydrolysis of the β-lactam bond, thus inactivating the β-lactam antibiotics for penicillins and cephalosporins. There are many beta-lactamases. Some are efficient at hydrolysing penicillins, some at hydrolysing cephalosporins and some are indiscriminate [2].
Cephalosporin compounds were first isolated from cultures of Cephalosporium acremonium from a sewer in Sardinia in 1948 by Italian scientist Giuseppe Brotzu. ESBLs are mainly derived from the ubiquitous TEM1/2, SHV-1 and CTX-M plasmid-mediated enzymes, which hydrolyse expanded spectrum cephalosporins to varying degrees. ESBLs are widespread all over the world, but the prevalence and phenotypic characteristics among clinical isolates may vary between geographical areas. Several studies in Kenya have noted an increase in resistance to third generation cephalosporins such as cefotaxime, ceftriaxone and ceftazidime in their Klebsiella isolates. This has partly been attributed to the production of extended spectrum β-lactamase enzymes by some of these bacteria. For instance, the reported existence of an extended-spectrum β-lactamase producing Klebsiella pneumoniae at the Kenyatta National Hospital and the observation of resistance to ceftriaxone [3]. The emergence of extended spectrum β-lactamase strains as potential pathogens requires careful screening to ensure accurate identification of these organisms and appropriate reporting of resistance to the physicians who are prescribing treatment for these patientsThis study aimed at determining which of the multi-drug resistant Klebsiella Pneumoniae isolates that showed resistance to the third generation cephalosporins, were Extended Spectrum β lactamases producers.
2. Materials and Methods
2.1. Bacterial Isolates
A total of 80 multi drug resistant clinical isolates of Klebsiella pneumoniae were obtained from two sources. The first source of the isolates was a previous study on anaerobes associated with Pelvic Inflammatory disease (P.I.D), KEMRI S.S.C No.495, while the other isolates were obtained from Kenyatta National Hospital medical microbiology laboratory. Isolations were done by routine laboratory diagnosis of patients with pneumonia, Urinary Tract Infections, burns and wound infections. Urine, blood, cerebrospinal fluid, sputum, stool and pus, high vaginal and throat swabs were examined for the presence of Klebsiella species at the routine Microbiology Laboratory of the Department of Medical Microbiology, University of Nairobi at the Kenyatta National Hospital. The isolates were identified by standard microbiological procedures including colony morphology and biochemical reactions. These biochemical tests were done alongside the use of API 20 E kit (La Balme les Grotles, Montalieu, Vercieu, France) for identification of Klebsiella pneumoniae subspecies pneumoniae.
2.2. Susceptibility Testing and ESBL Production
A McFarland nephelometer was used to adjust the turbidity of bacteria suspensions to predetermined levels. For bacterial susceptibility tests, the 0.5 McFarland turbidity standard corresponding to approximately 106 colony forming units per millilitre of bacterial cell suspension was used. The sensitivity of isolates to third-generation cephalosporins (3GCs), ceftazidime (30 μg), cefotaxime (30 μg), ceftriaxone (30 μg) and to other antibiotics such as ampicillin (10 μg), amikacin (30 μg), aztreonam (10 µg), cefuroxime (30 μg), cefoxitin (30 μg), gentamicin (10 μg), imipenem (10 μg), ciprofloxacin (30 μg), cefepime (30 μg), chloramphenicol (10 µg), ciprofloxacin (5 µg), cotrimoxazole (30 μg), kanamycin (10 µg), augmentin®, piperracillin (10 µg), streptomycin (10 µg), sulphamethoxazole (30 μg), tetracycline (10 µg) and amoxycillin (10 µg) (HiMedia, Mumbai, India) was performed by disc diffusion method on Mueller Hinton agar plates. The diameter of the zone of inhibition for each antibiotic was measured and interpreted as resistant, intermediate susceptible or susceptible according to NCCLS criteria [4]. The minimum inhibitory concentration of the antimicrobial agents was assessed against 80 Klebsiella pneumoniae isolates using the E test strips. Breakpoint MIC values were interpreted as recommended by the National Committee for Clinical Laboratory Standards [4]. Isolates that showed intermediate or complete resistance to any of the 3GCs were selected for further ESBL detection by double disk diffusion synergy test (DDST) to determine synergy between a disk of coamoxyclav that is, augmentin® (20 µg amoxycillin and 10 µg clavulanic acid) and 30 µg disk of each of the 3GC antibiotics placed 20 mm centre to centre from augmentin® disc. E. coli ATCC (American Type Culture Collection) 25922 was used as the negative control and Klebsiella pneumoniae ATCC 700603 was used as the positive control for ESBL production. The test organism was considered to produce ESBL if the zone size around the test disc increased towards the co-amoxyclav disc or if neither disks were inhibitory alone but bacterial growth was inhibited where the two antibiotics diffused together.
3. Results
ESBL production was detected in 30 (37.5%) isolates. All the isolates were susceptible to imipenem (Table 1). When the isolates were subjected to susceptibility testing, resistance to ampicillin was the most prevalent among 70 (87.5%) isolates followed by resistance to piperracillin.
The interpretation of the susceptibility profiles of the thirty ESBL-producing isolates of Klebsiella pneumoniae to 30 μg disks of cefotaxime, ceftazidime and ceftriaxone are shown in Table 2. The distribution of zone diameters did not differ between strains able or not able to transfer ESBL production. It is evident that many ESBL producing isolates appeared to be susceptible by disk diffusion method. Of the two methods which were used for detection of ESBL production in this study, phenotypic confirmatory disc diffusion test (PCDDT) was a more sensitive procedure for detection of ESBL than the DDST. Twenty seven (90%) of the 30 ESBL producing strains
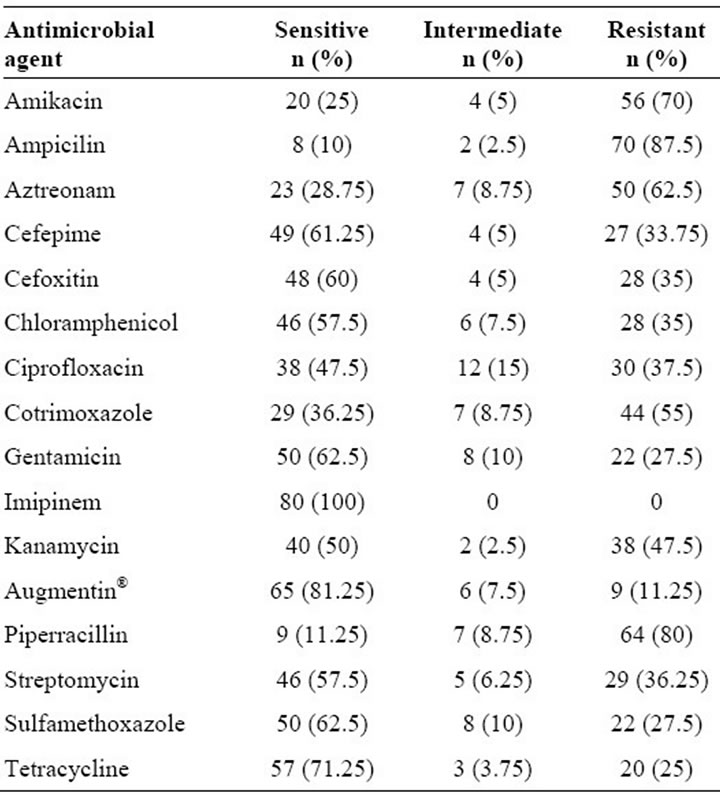
Table 1. Susceptibility profiles of all the Klebsiella pneumoniae isolates in the study by the disk diffusion technique.

Table 2. Susceptibility of ESBL producing K. pneumoniae isolates to third generation cephalosporins (n = 30).
were detected by DDST using cefotaxime, ceftriaxone and ceftazidime antibiotics. Using standard double disk synergy test (DDST) as screening method for identifying potential ESBL producers, ceftriaxone was the most efficient antimicrobial in screening isolates as potential ESBL producers followed by cefotaxime. In this test, a disk containing amoxicillin-clavulanate (AMC) was placed in proximity to disks containing ceftazidime, cefotaxime and ceftriaxone antibiotics. The results showed that the clavulanate in the amoxicillin-clavulanate disk diffused through the agar and inhibited the β-lactamase surrounding the cefotaxime and ceftriaxone antibiotics disks. Enhancement of the inhibition zone of any of the third generation cephalosporins tested, on the side facing the amoxicillinclavulanate disk was interpreted as a positive test. Ceftriaxone missed three ESBL harbouring isolates. Cefotaxime and aztreonam missed seven and nine ESBL harbouring isolates respectively. Ceftazidime was the least efficient among these four antimicrobials missing fourteen isolates that were later identified as harbouring ESBLs.
ESBLs derived from TEM and SHV β-lactamases do not provide resistance to cefoxitin. Table 2 shows that 24 of the 30 ESBL-producing isolates of Klebsiella pneumoniae were resistant to ceftazidime by disk diffusion test, while 18 were resistant to Cefotaxime and Ceftriazone. These results suggest that resistance to these cephalosporins is largely attributed to production of ESBL. Among the isolates shown not to produce ESBL, eleven of them were sensitive to cefotaxime (Table 3). However, there was a significant lower number of sensitive ESBL-producing isolates to the third generation cephalosporins as compared to the sensitive isolates that were not ESBL producers. Such results have significant implications in clinical practice.
4. Discussion
In the present study, resistance patterns from both disk diffusion tests and the determination of minimum inhibitory concentrations showed resistance to most of the drugs tested. Disk diffusion results showed that all the isolates were sensitive to imipinem. Among all the eighty isolates for which MICs were determined, 68 (85%) isolates were resistant to amoxicillin, but tests with Augmentin® (amoxicillin-clavulanic acid) showed resistance in only 10 (12.5%) of the isolates. The higher activity of Augmentin® is attributable to inhibition of β-lactamases by the clavulanic acid in the amoxicillin-clavulanic acid combination. A comparison between the phenotypic confirmatory disk diffusion method and double-disk synergy test (ceftazidime, ceftriaxone, cefotaxime, and amoxicillin/clavulanic acid) was made to assign the appropriate method of detection for ESBLs. The double disk synergy test (DDST) was found to be a useful, simple and cost effective test for the detection of ESBL producing strains. The results indicated that thirty out of the eighty klebsiellae pneumoniae isolates in the present study were producing Extended spectrum β-lactamases. This is relatively a high prevalence (37.5%) of ESBL production. Extended spectrum β-lactamases appear to have been a major cause of the resistance encountered in the Klebsiella isolates studied. A similar study also showed that Klebsiellae organisms sometimes exhibit multiple resistance [5]. The emergence and spread of Klebsiella pneumoniae resistance to third generation cephalosporins has been associated with the over use of these agents especially in intensive care units [6].

Table 3. Susceptibility of ESBL non-producing K. pneumoniae isolates to third generation cephalosporins (n = 50).
Although enzymatic destruction is the most common mechanism of resistance to the β-lactam antibiotics, this may be accompanied by reduced accumulation of drugs in the cell and there could as well be an alteration of penicillin-binding-proteins which are the targets of these antibiotics. Extended spectrum β-lactams are commonly included in the empirical antibiotic regimens for treatment of gram negative sepsis. The increasing use of broad spectrum cephalosporins has become one of the major factors responsible for the high rate of selection of extended spectrum beta lactamase producing microorganisms [7]. Resistant strains are not inhibited by the usually achievable systemic concentrations of the agent with normal dosage schedules and/or fall in the range where specific microbial resistance mechanisms are likely (for example, β-lactamases) and clinical efficacy has not been reliable in treatment studies. These results show that most of the Klebsiella isolates dealt with in this study exhibited multiple resistance to the antimicrobial agents tested. The results have significant implications. It has been reported that many Klebsiella pneumoniae strains that produce extended spectrum β-lactamases also show associated resistance to other antimicrobial agents [8]. The risk of treatment failure is a reality especially while treating infections caused by ESBL-producing bacteria with third generation cephalosporins since some organisms, for instance some isolates in this study, would otherwise be reported as susceptible to antibiotics by routine antimicrobial susceptibility testing. The real challenge is the ESBL producing organisms for which MICs of 3GC are in the susceptible range and they may not be truly susceptible when serious infections are considered although these isolates may have been reported susceptible. In that regard, treatment recommendations for infections caused by organisms producing ESBL’s currently include carbapenems in the first-line therapy and ciprofloxacin and β-lactam/β-lactamase inhibitor combinations as the second-line therapy. It is very important for the infectious diseases practioner to be sure that the clinical microbiology laboratory is capable of detecting these important beta-lactamase enzymes. This is despite the fact that ESBL detection is difficult and remains a challenge for automated susceptibility testing systems.
In conclusion, further studies are needed to establish the optimal technique for detecting ESBL production. Meanwhile, it should be recognized that the present disk diffusion criteria underestimates the prevalence of the ESBL producing strains.
NOTES