Horizontal gene transfer of plant-specific leucine-rich repeats between plants and bacteria ()
1. INTRODUCTION
LRRs (Leucine Rich Repeats) are present in 20,727 proteins in the PFAM database [1], 14,316 in SMART [2], 20,937 in PROSITE [3], and 29,365 in InterPro [4]. The repeat number of tandem LRRs ranges from two to sixty-two. LRR-containing proteins have been identified in viruses, bacteria, archaea, and eukaryotes. Most living organisms have at least one LRR protein, so far as we know. Most LRR proteins are involved in protein, ligand in protein, protein interactions; these include the plant immune response and the mammalian innate immune response [5-10].
All LRRs can be divided into a HCS (highly conserved segment) and a VS (variable segment). The HCS part consists of an eleven residue stretch, LxxLxLxxNxL, or a twelve residue stretch, LxxLxLxxCxxL, in which “L” is Leu, Ile, Val, or Phe, “N” is Asn, Thr, Ser, or Cys, and “C” is Cys, Ser or Asn. Three residues at positions 3 to 5, xLx, form a short b-strand. These b-strands from tandem LRRs stack parallel and the LRRs then form an LRR arc or domain. The concave face of the arc consists of a parallel b-sheet (or rarely an anti-parallel b-sheet) whose strands are approximately parallel to the axis of the arc. The convex face is made of a variety of secondary structures including the a-helix, 310-helix, polyproline II helix, and an extended structure or a tandem arrangement of b-turns. In most LRR arcs the b-strands on the concave surface and (mostly) helical elements on the convex surface are connected by short loops or bturns. Most of the known LRR domains have a cap that shields the hydrophobic core of the first LRR unit at the N-terminus (N-cap) and/or the last unit at the C-terminus (C-cap). In extracellular proteins or extracellular regions, these caps frequently consist of Cys clusters consisting of two or four Cys residues; the Cys clusters on the Nand C-terminal sides of the LRR arcs are called LRRNT and LRRCT, respectively [7,11].
Eight classes of LRRs have been recognized; they are characterized by different lengths and consensus sequences of the VS part of the repeats [12]. They are “RI-like”, “CC”, “Bacterial”, “SDS22-like”, “plant specific (PS)”, “Typical”, “TpLRR”, and ”IRREKO” [6, 12,13]. GALA-LRR was proposed as a subclass of CC-LRR [14]. These classes of LRR domains adopt a variety of similar structures. Correspondingly, the several LRR domains form LRR arcs of varying diameters. Matsushima et al. [15,16] reported that LRR domains consisting of two different LRR classes of “Typical” and “Bacterial” are present in eukaryotic LRR proteins such as the subfamily of small LRR proteoglycan (SLRP), and the subfamily of Toll-like receptors (TLR7, TLR8 and TLR9). Moreover, “IRREKO” and “SDS22-like” or “Bacterial”, and “GALA” and “CC” coexist in bacterial LRR proteins [13,14].
The consensus sequence of PS-LRRs is LxxLxLxxNxLsGxIPxxLGxLxx in which uppercase and lowercase indicates more than 60% and 20% identity, respectively [14]; “L” is Leu, Val, or Ile, “N” is Asn, Cys, Ser, Thr, “s” is Ser, “G” is Gly, “I” is Ile or Leu, “P” is Pro, and “x” is a non-conserved residue. The repeat length is 23 - 25 residues. PS-LRR domains are observed in many plant proteins [12,14]; these include LRR-containing receptor-like kinase proteins (LRR-RLKs) [17], LRR-containing receptor-like proteins (LRR-RLPs) [18], and polygalacturonase inhibiting proteins (PGIPs) [19], which are involved in disease resistances and/or development. LRRRLKs have an extracellular LRR domain with an N-terminal signal peptide, a single trans-membrane spanning region and an intracellular serine, threonine kinase region. LRR-RLPs have a short cytoplasmic tail instead of the kinase region.
Crystal structures of the PS-LRR domain are available for Phaseolus vulgaris polygalacturonase-inhibiting protein (PGIP) and Arabidopsis thaliana LRR-RLK BRI1 [20-22]. PGIP contains ten PS-LRRs most of which are 24 residues long. On the convex side, nine 310-helices are almost parallel to the b-strands on the concave side. The consensus sequence LxGxIP at positions 11 to 16 likely forms a second b-strand that characterizes the fold of the PS-LRRs. Thus, structural units of the PS-LRRs may be represented as b-b-310 [7]. PGIPs have both an LRRNT with Cx29CCx8C forming two disulfide bonds and an LRRCT with Cx21CxCx6C also forming two disulfide bonds. A similar structural feature also exists in A. thaliana BRI1 [21,22].
“Typical” LRRs are the most abundant LRR class [6,12]. The consensus sequence is LxxLxLxxNxLxxLpxxoFxxLxx in which uppercase indicates more than 50% occurrence of a given residue in a certain position; lowercase indicates 30% - 50% occurrence; “L” is Leu, “N” is Asn, “p” is Pro, “o” indicates a non-polar residue, “F” are Phe, and “x” is a non-conserved residue [6,12]. The repeat length is 20 - 27 residues. Their variable segments adopt mainly polyproline II plus b-turn, consecutive b-turns, or b-turn plus polyproline II conformations in the convex faces; the structural units may be represented by b-(b-turn + polyproline II) [23]. “RI-like” LRRs are contained in proteins such as ribonuclease inhibitor [6,12]. The consensus sequence is LxxLxLxxNx- (L/C)xxxgoxxLxxoLxxxxx. The repeat length is 28 - 29. Most of their VSs adopt a-helical conformations [23]. Cysteine containing (CC) LRR proteins include Saccharomyces cerevisiae GRR1 protein. The consensus sequence is LxxLxLxxCxxITDxxoxxL(a/g)xx(C/L)xx [6, 12]. The repeat length is 25 - 27. Most of their VSs adopt a-helical conformations [23]. GALA-LRR is a subclass of CC-LRR; its consensus sequence is LxxLxLxxNxIgdx(g/ a)OxxLax(n/s/d)xx of 24 residues [14]. “SDS22-like” LRRs are included in SDS22, internalin-A, and internalin-B [6,12]. The consensus sequence is LxxLxLxxN(r/ k)I(r/k)(r/k)IE(N/G)LExLxx. The repeat length is 21 - 23. The individual units are in b-310 conformation [23]. “Bacterial” LRRs are found in Yersinia pestis YopM, and in Shigella flexneri IpaH. The consensus sequence is LxxLxVxxNxLxxLP(D/E)LPxx [6,12]. The repeat length is 20 - 22. The structural units are in b-polyproline II conformations [23]. “TpLRR” is found in Treponema pallidum LRR protein and in Bacteroides forsythus surface antigen. The consensus sequence is LxxLxLxxxLxxIgxxAFxx(C/N)xx [6,12]. The repeat length is 23 - 25. The dominant feature is a highly conserved segment of ten residues, differing from the corresponding eleven residues of other LRRs. The structure of this class remains unknown. “IRREKO” are found in many bacterial proteins including internalin-J [13]. The consensus sequence is LxxLxLxxNxLxxLDLxx(N/L/Q/x) xx or LxxLxCxxNxLxxLDLxx(N/L/x)xx. This class is characterized by a nested periodicity; it consists of alternating 10- and 11-residues units of LxxLxLxxNx(x/-). The structural units are in b-extended conformations [24].
The evolution of LRRs is not well understood. It is not even known whether all LRR’s share a common ancestor. Kobe and Deisenhofer [5] pointed out the possibility of their having been at least a few independent origins of LRRs. Kajava noted that an LRR domain never contains mixtures of different types of LRR repeats and suggested separate origins for the different classes of LRRs [12,14]. In contrast, Andrade et al. [25] suggested that LRRs have a common origin. Matsushima et al. [13,15,16] proposed that the four LRR classes of “Bacterial”, “Typical”, “SDS22-like” and “IRREKO” might have evolved from a common ancestor.
The evolution of plant disease resistance (R) genes that encode LRR domains has been studied by many researchers. The generation of the genes that encode entire LRR proteins has been proposed to involve gene duplication and fusion, genetic recombination, diversifying selection, and sequence divergence in the inter-genic region as well as in the composition of the transposable elements [26]. The possibility of horizontal gene transfer (HGT) of proteins containing “TpLRR”, GALA-LRR or some other LRR has been discussed [14,27,28]. HGTs of some genes from plants to an animal—Elysia chlorotica —or an opisthokonta—Adineta vaga—have also been reported [29].
PS-LRRs had previously been recognized only in plant proteins; most (or all) plants have at least one PS-LRR protein. However, we find that some proteins from bacteria contain PS-LRR domains. The focus of this paper is to investigate the origin of bacterial PS-LRR domains. Here we document the occurrence of a PS-LRR domain in 20 proteins from eleven bacterial species in three phyla. Analyses of the distribution of organisms having PS-LRR proteins and of similarity searches of both amino acids sequences and nucleotides sequences, as well as results of the program EGID (Ensemble algorithm for Genomic Island Detection), indicate that HGT event(s) of genes/gene fragments encoding PS-LRR domains occurred between bacteria and plants, and HGT among the eleven bacterial species, or the three phyla, as opposed to descent from a common ancestor. There is the possibility of the occurrence of a single HGT event from plant to bacteria.
2. MATERIALS AND METHODS
2.1. Database Similarity Search
We recently developed a new method (LRRpred) that utilizes known LRR repeats to recognize and align new LRRs [16,30]. LRRpred incorporates multiple sequence alignments and secondary structure predictions. It predicts correctly the number of LRRs, their lengths and their boundaries. First, we selected regions containing canonical tandem PS-LRRs in plant proteins such as tomato Cf-2 using LRRpred. Second, we performed sequence similarity searches using the amino acid sequences of all PS-LRRs within the tandem domain as queries in FASTA [31] at the Bioinformatic Center, Institute for Chemical Research, Kyoto University on December 22, 2009 (http://www.genome.jp/tools/fasta/). These searches identified bacterial proteins having PSLRRs. Third, we confirmed PS-LRRs in these detected bacterial proteins by LRRpred. Fourth, we performed sequence similarity searches using both amino acid sequence and nucleotide sequence of bacterial PS-LRRs as queries by FASTA and then considered eukaryotic proteins under the following conditions as putative homologs. The database searches using the nucleotide sequence show highly significant similarity with E-value < 10−10 and their overlapping length is larger than 70% of the query nucleotide length [32]. The database searches using the amino acid sequence of LRRs in Dalk_4722 show highly significant similarity with E-value < 10−20. LRR proteins from Leishmania major strain Friedlin, L. braziliensis MHOM/BR/75/M2904, and L. donovani were also collected by use of keywords in the NCBI database. All LRRs including PS-LRRs in the LRR proteins were identified by LRRpred. A similarity network of nucleotide sequence of PS-LRRs in bacterial PS-LRR proteins and eukaryotic PS-LRR proteins identified here was drawn by Cytoscape (version 2.8) [33].
2.2. Sequence Analyses
The protein localization sites in cells were predicted by PSORT [34]. Signal sequence analysis was carried out using the multiple programs of SignalP [35], SIG-Pred (http://bmbpcu36.leeds.ac.uk/prot_analysis/Signal.html), Signal-3L [36], and PrediSi [37]. If some sequence was preferred by any one of the four programs, the sequence was identified as a signal peptide. Transmembrane predictions were produced by TMHMM. Except for signal peptides and transmembrane regions in PS-LRR proteins, other characteristic regions were identified using PFAM and/or SMART. The consensus sequence of PS-LRRs was determined by WebLogo [38].
2.3. EGID Analysis
Genomic islands are regions of the genome that were originally transferred from other organisms. Ensemble algorithm for Genomic Island Detection (EGID) utilizes the prediction results of existing computational tools, then filters and generates consensus prediction results [39]; their computational tools are AlienHunter [40], Centroid [41], COLOMBO SIGI-HMM [42], IslandPath [43], INDeGenIUS [44], and PAI-IDA [45].
3. RESULTS
3.1. Bacterial Proteins Having PS-LRR Domains
Database searches using the amino acid sequences of PS-LRRs in plant proteins, such as tomato Cf-9, detected 20 proteins from eleven bacterial species that have PSLRR domains (Table 1, Figure 1, and Appendix 1); the PS-LRRs in tomato Cf-9 protein show good matches to the consensus of LxxLxLxxNxLxGxIPxxLxxLxx [46]. These 20 PS-LRR proteins consist of seven proteins from Proteobacteria (Desulfatibacillum alkenivorans and Beggiatoa sp. PS.), three from Cyanobacteria (Synechococcus sp. and Crocosphaera watsonii), and ten from Bacteroidetes (Flavobacterium johnsoniae, Leeuwenhoekiella blandensis, Dokdonia donghaensis, Robiginitalea biformata, Flavobacteriales bacterium, and Bacteroides coprocola) [47-55]. The entire genomes of thirty-one cyanobacterial species have been determined. However, only two cyanobacterial species contain PS-LRR proteins. Most of these organisms are found in marine or water environments; while only B. coprocola is found in human faeces (Table 1).

Table 1. Bacterial proteins having a PS-LRR domain.

Figure 1 . Schematic representation of twenty bacterial proteins having PS-LRR repeats from eleven species. Desulfatibacillum alkenivorans Dalk_4722; Beggiatoa sp. PS. BGP_2706; Beggiatoa sp. PS. BGP_2932; Beggiatoa sp. PS. BGP_1054; Beggiatoa sp. PS. BGP_0730; Beggiatoa sp. PS. BGP_4203; Beggiatoa sp. PS. BGP_0049; Synechococcus sp. CYB_1422; Synechococcus sp. CYA_1022; Crocosphaera watsonii. CwatDRAFT_2187; Flavobacterium johnsonia Fjoh_0602; F. johnsonia Fjoh_1865; Leeuwenhoekiella blandensis MED217_10857; Dokdonia donghaensis MED134_03679; Robiginitalea biformata RB2501_09035; Flavobacteriales bacterium FB2170_08714; F. bacterium FB2170_07319; F. bacterium FBALC1_03992; Bacteroides coprocola BACCOP_00862; B. coprocola BACCOP_03537.
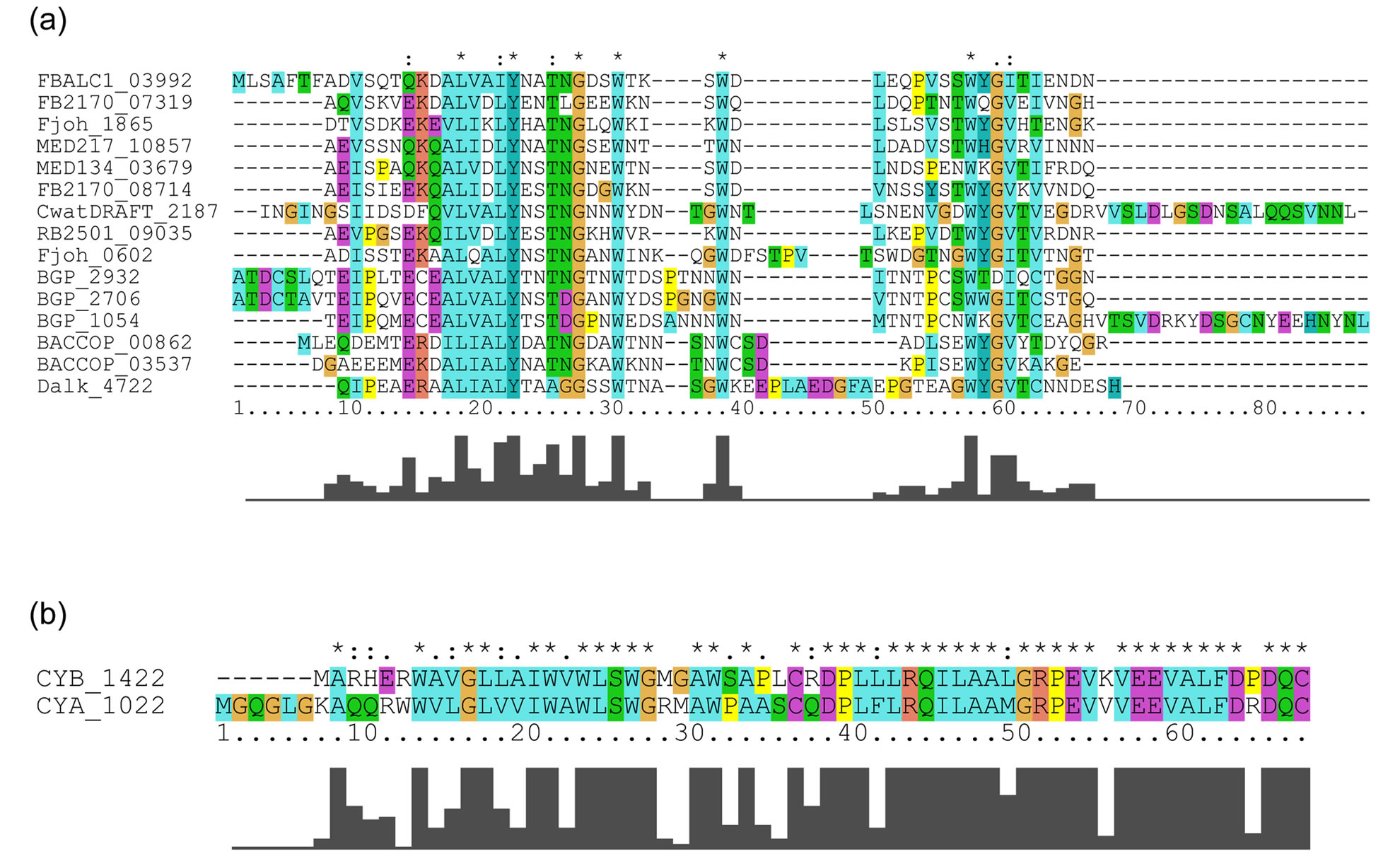
LRRs in the 20 bacterial proteins belong to the PS-LRR class. The VS part of the C-terminal LRR in some LRR domains does not honor this consensus. A similar situation is frequently observed in other LRR classes [7,9]. The WebLogo outputs show the occurrence frequency of amino acids at each position (Figure 2). The consensus sequence of the LRRs is LexLxLsnNqLsGs(I/l)Px(e/s)(i/l)gnLtn in which “L” or “l” is Leu, “I” or “i” is Ile, “N” or “n” is Asn, “s” is Ser, “G” or “g” is Gly, “e” is Glu, “q” is Gln, “P” is Pro, “t” is Thr, and “x” is a non-conserved residue; uppercase indicates more than 60% occurrence of a given residue in a certain position;
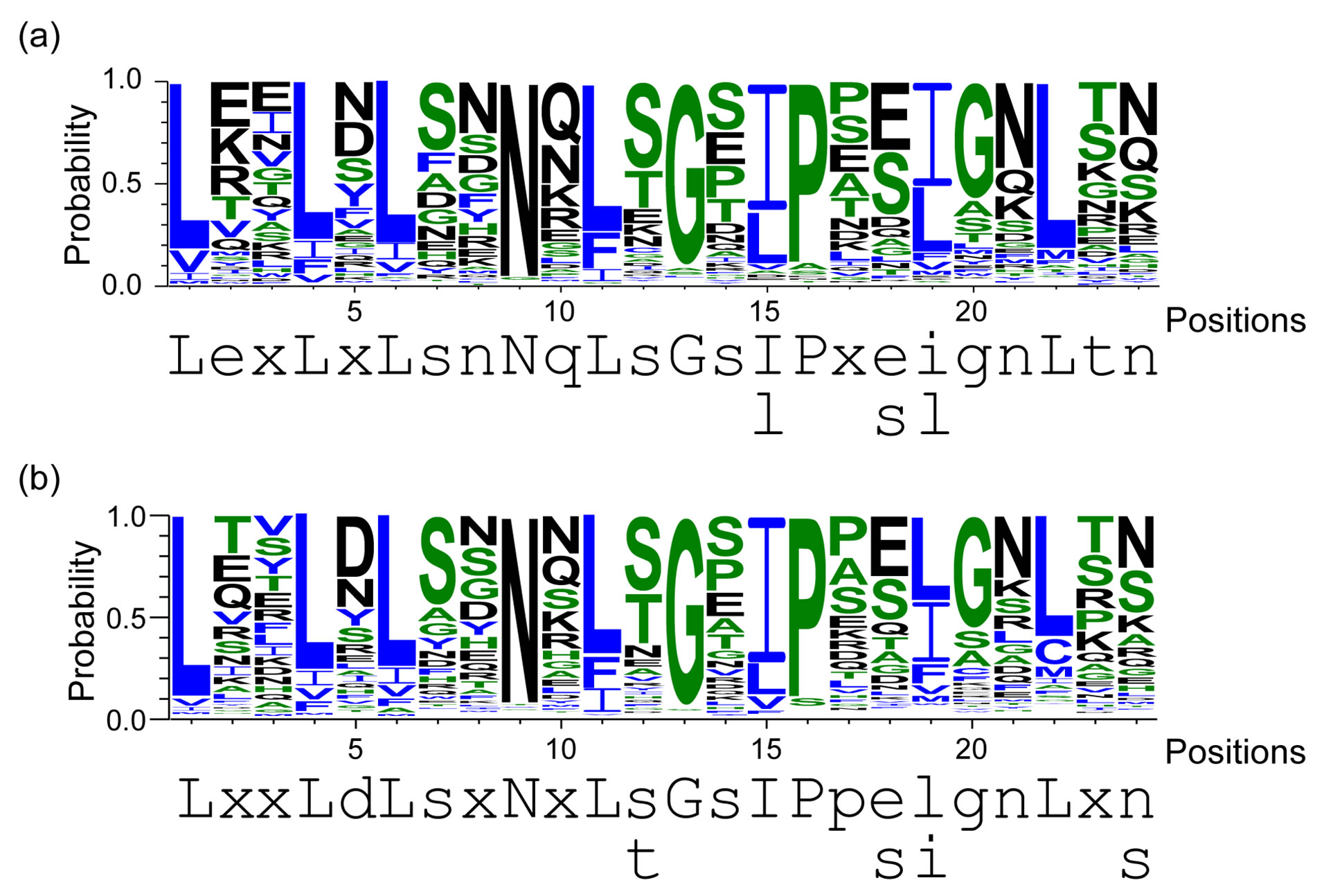
Figure 2. The overall consensus sequences of the PS-LRR repeats in bacterial PS-LRR proteins and in plant PS-LRR proteins. (a) Overall consensus sequences of bacterial PS-LRRs from a total of 159 LRRs in twenty bacterial PS-LRR proteins; (b) Overall consensus sequences of eukaryotic PS-LRRs from a total of 1863 LRRs in the 83 eukaryotic PS-LRR proteins identified here. The upper and the lower portions are WebLogo output and the consensus sequence of PS-LRRs, respectively. Uppercase indicates more than 60% occurrence of a given residue in a certain position; lowercase indicates 20% - 60% occurrence.
lowercase indicates 20% - 60% (Figure 2(a)).
This bacterial LRR consensus shows the most similarity with the PS-LRR consensus, because the conserved residues, conserved hydrophobic patterns, and repeat lengths are almost identical, and the occurrence frequencies of the consensus residues completely satisfy the criterion of Kajava [14]. Most important, the PS-LRRs are characterized by the consensus sequence LxGxIP at positions 11 to 16 that forms a second b-strand; this characteristic sequence is not seen in the other seven classes at all. Moreover, Gly at the 13th position is almost completely conserved with 90% occurrence (Figure 2(a)). Thus, this bacterial PS-LRR consensus differs from the LRR consensus of the other classes of “RI-like”, “Typical”, “Bacterial”, “SDS22-like”, “TpLRR”, ”IRREKO”, and “CC”/“GALA” [12,14,56]. Five classes— “Bacterial”, “SDS22-like”, “TpLRR”, “IRREKO”, and “GALA”— have been recognized in bacterial LRR proteins. There are other LRR motifs in proteins from bacteria and virus [28,57,58]. However, these other LRR motifs clearly differ from the bacterial PS-LRR.
The repeat number of tandem PS-LRRs from bacteria ranges from 2 to 15; F. johnsoniae Fjoh_0602 has 15 LRRs; D. alkenivorans Dalk_4722 has nine PS-LRRs, all of which are 24 residues long (Figure 1). Six PS-LRR proteins from Beggiatoa sp. PS. contain 3 to 14 PS-LRRs (Figure 1), which are sometimes variable in length. The PS-LRR domains from Synechococcus sp. CYA_1022 and CYB_1422 are orthologous; the sequences of their full lengths are 81.3% identical and the LRR sequences with 217 residues are 87.1% identical. BGP_4203 and BGP_0730 both have a single transmembrane helix and are paralogous. The full length of BGP_4203 with 70 residues is 68.6% identical to the C-terminal, 80 residues of BGP_0730 with 232 residues.
BGP_2706 and BGP_2932 both have an LRRNT with Cx10C29Cx6C. BGP_1054 has an LRRNT with
Cx10C29Cx6Cx15C and an LRRCT with Cx25C. CYA_1022 and CYB_1422 have an LRRNT with Cx30C and an LRRCT with Cx22C. The putative N-cap regions in the bacterial PS-LRR proteins contain a conserved motif of Lx8Wx2-13Wx5-10Wx1GV (Appendix 2); while those of CYA_1022 and CYB_1422 have a different conserved motif and are more closely related to putative N-cap regions in some plant PS-LRR proteins. The former conserved motif is seen in a region including the LRRNT of PGIP and the Trp contributes to the hydrophobic core of the N-cap structure [20]. Seventeen of the bacterial PSLRR proteins have putative N-cap/LRRNT regions (Figure 1).
Five of these twenty bacterial proteins contain domains in addition to the LRR domains (Figure 1). BGP_1054 contains one PKD (Polycystic Kidney Disease) domain [59] at the C-terminal side (Figure 1). CwatDRAFT_2187 contains a Calx-b domain [60] at the N-terminal side of the PS-LRR domain and a putative immunoglobulin domain [61] on its C-terminal side. Fjoh_0602 contains a single immunoglobulin domain and twelve repeats of PKD at the C-terminal side. BACCOP_00862 and BACCOP_03537 contain both an AhpC-TSA family domain [62] and/or an IgA peptidase M64 family domain [63] at the C-terminal side. Three BGP proteins contain trans-membrane spanning regions. PSORT [34] predicted that sixteen of the 20 bacterial LRR proteins are extracellular; CwatDRAFT_2187 is in the outer-membrane region and BGP_0049 is in the cytoplasmic-membrane region, while locations of BGP_ 4203 and Fjoh_1865 are unknown (Table 1).
3.2. Eukaryotic Proteins Having a Bacterial PS-LRR Domain
Database similarity searches by FASTA [31] using the amino acid sequences of all PS-LRR domains in the 20 bacterial proteins were done. The database search using nine PS-LRRs in Dalk_4722 with 216 residues found a high degree of identity with 495 eukaryotic proteins as well as with three bacterial proteins (Fjoh_0602, CYA_1022, and CYB_1422) with E-values < 10−20. The pair wise comparisons between Dalk_4722 and the eukaryotic proteins show 32.7% - 46.9% identity in 182 - 254 residue overlap. The greatest similarity is seen in A. thaliana LRR-RLK having 24 PS-LRRs [AT1G34110] with E-value = 4.0 × 10−34 (Figure 3); the pair wise comparison shows 43.9% identity in a 214 residue over-
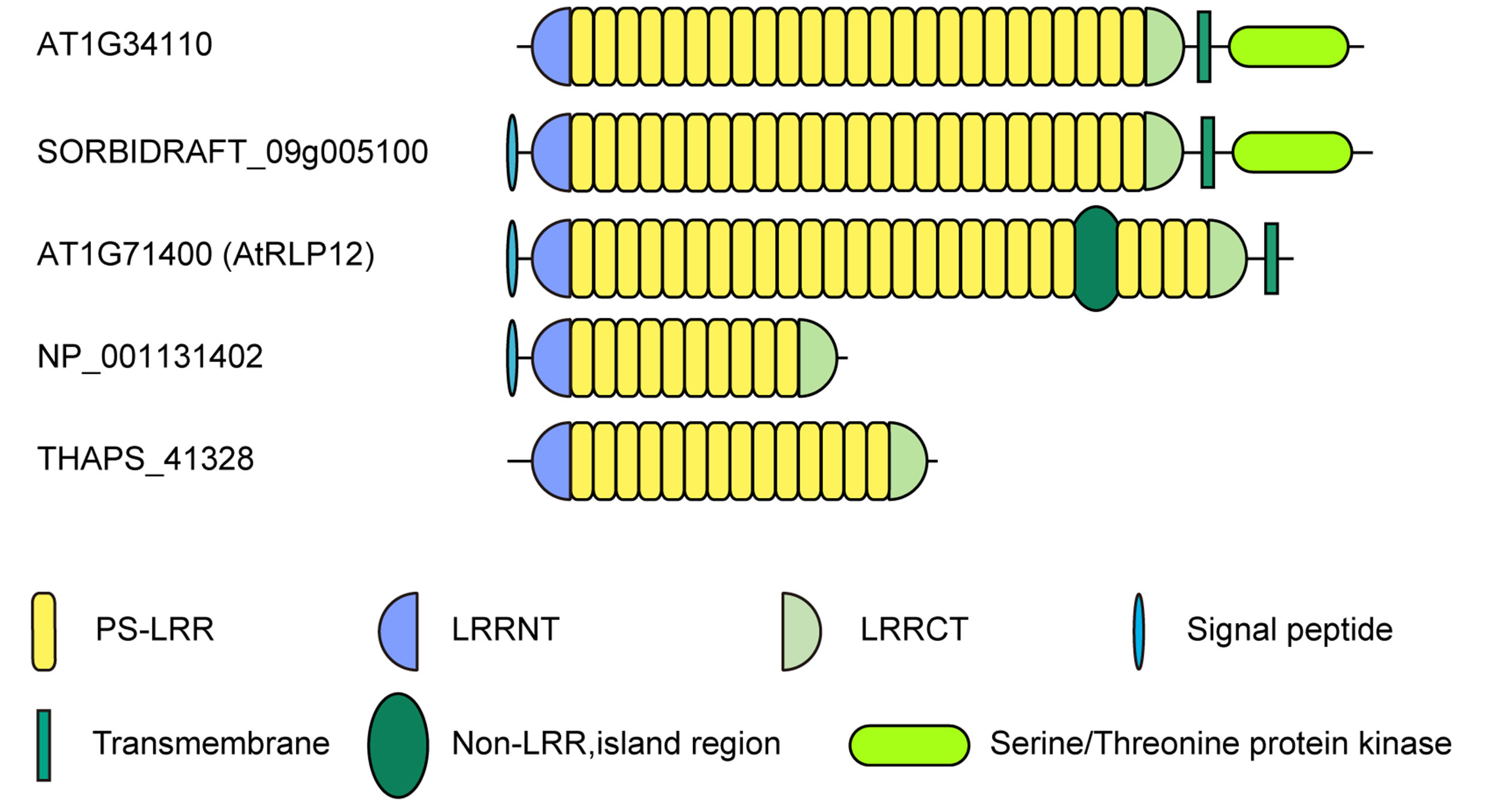
Figure 3. Schematic representation of eukaryotic PS-LRR proteins. LRR-RLK, Arabidopsis thaliana AT1G34110 [NP_174673] and Sorghum bicolor SORBIDRAFT_09g005100 [XM_002439314]; LRR-RLP, Arabidopsis thaliana AtRLP12 [AT1G71400]; Extracellular PS-LRR protein; Zea mays hypothetical protein [NP_ 001131402]; Thalassiosira pseudonana THAPS_41328 [XM_002296129].
lap. The 495 eukaryotic PS-LRR proteins have 10 to 35 PS-LRRs in their domains. Similar results were observed using other bacterial PS-LRR domains as probes.
As noted, we employed a cut-off E-value of 10−20 as a criterion of highly significant similarity to bacterial PSLRRs. Many proteins with 10−4 > E-values > 10−20 have other LRR motifs with the consensus of LxxLxLxxNxLxxLPxxLGxLxx, LxxLxLxx(C/N)xxLxxLPxxLGxLxx or LxxLxLxxNxL(T/S)GxIPxxWxx(M/L)xx. For example, MICPUN_84219 with significant similarity (E-value = 4.7 × 10−19) contains 14 LRRs with the consensus of LxxLxLxxNxLTSVPAEIGQLTS of 23 residues. These LRR motifs are not regarded as PS-LRR.
The database similarity searches using the nucleotide sequences coding LRRs in Dalk_4722 detected two plant PS-LRR proteins with E-values < 10−10. This similarity search did not identify the other LRR motifs. Similar results were observed using the nucleotide sequences of other bacterial PS-LRR domains as probes. For putative homologs of PS-LRR domains having a variety of LRR repeats within different bacterial proteins, we employed database similarity searches using the nucleotide sequences; although, amino acid sequences searches are more sensitive to identification of homology. We considered eukaryotic proteins with the following conditions as putative homologs. The PS-LRRs have highly significant similarity with E-value < 10−10 and their overlapping length is larger than 70% of the query nucleotide length. They are a subset of the whole set of PS-LRR homologs.
The database searches using bacterial PS-LRRs as probes found a high degree of similarity with 83 proteins from eight eukaryotic species (Figure 3 and Appendix 3). These 83 eukaryotic PS-LRR proteins are from monocots (rice—Oryza sativa, Zea mays, and Sorghum bicolor), dicots (A. thaliana, poplar—Populus trichocarpa, and grape—Vitis vinifera), and a bryophyte (moss—Physcomitrella patens), as well as from one diatom—Thalassiosira pseudona. No archaeal PS-LRR proteins were identified.
The 83 eukaryotic PS-LRR proteins have seven to forty-three PS-LRRs in their domains. The PS-LRR consensus is LxxLdLsxNxL(s/t)GsIPp(e/s)(i/l)gnLx(n/s) (Figure 2(b)). The comparison of the consensus sequences between bacterial and eukaryotic PS-LRRs demonstrates that positions 1, 4, 6, 9, 11, 13, 15, 16, and 22 are mainly occupied by respective conserved residues. In addition, a significant, weak conservation is observed at positions 5, 7, 12, 14, 17, 18, 19, 20, 21, and 24. Thus, bacterial PS-LRRs are significantly more similar to eukaryotic PS-LRRs than to the other seven classes.
The PS-LRR domains frequently have both an LRRNT and an LRRCT (Figure 3), as well as PGIP and BRI1. The plant proteins are mostly LRR-RLKs (75 of the 83 eukaryotic proteins) and the remaining are LRRRLPs or extracellular proteins that have a signal peptide (but have no transmembrane helix) (Figure 3). There are 239, 357, and 440 LRR-RLKs from A. thaliana, O. sativa, and P. trichocarpa [64], a total of 1,036. The seventy-five identified LRR-RLKs are a part of the 1036 LRR-RLKs.
3.3. Striking Similarity of Nucleotide Sequence in Bacterial and Eukaryotic PS-LRRs
BGP_1054 has two PS-LRR domains; the second PS-LRR domain contains 14 repeats (Figure 1). A nucleotide similarity search using the 14 PS-LRRs identified a large number of eukaryotic proteins (42 of the 83 eukaryotic proteins) as well as two bacterial proteins
(Fjoh_0602 and BGP_0730) with highly significant similarity (E-values < 10−10); the pair wise comparisons show 51.7% - 60.2% identity in 724 - 973 nt overlap. The 14 units of the second BGP_1054 PS-LRR domain have the greatest similarity to POPTRDRAFT_586452, which has 22 PS-LRRs and is an LRR-RLK, with E-value = 2.5 × 10−35; the pair wise comparison shows 58.4% identity in a 973 nt overlap; this corresponds to most of the PS-LRR domain in POPTRDRAFT_586452. Despite a large evolutionary distance between the bacterium (Beggiatoa) and the plant (poplar), this high degree of identity is comparable to the 50% - 60% identity between seven internal exons, which encode two “RI-like” LRRs in ribonuclease inhibitor from human, pig, and mouse [65]. Consequently, the LRR repeats in the nine bacterial PS-LRR proteins show highly significant similarity with those in the 83 eukaryotic proteins.
A similarity network of nucleotide sequence of PSLRRs in both the 20 bacterial PS-LRR proteins and the 83 eukaryotic PS-LRR proteins is shown in Figure 4" target="_self"> Figure 4 [33]. As expected, CYA_1022 shows the most similarity with CYB_1422. In addition, these two proteins are more similar to eukaryotic PS-LRR proteins than to other bacterial PS-LRR proteins. Similarly, Dalk_4722, RB2501_ 09035, BACCOP_00862, and BACCOP_03537 are highly similar to only eukaryotic PS-LRR proteins. Some bacterial PS-LRR proteins are similar to those from different bacterial species or from different phyla; BGP_ 1054-Fjoh_0602, BGP_2932-Fjoh_0602, and MED134_ 03678-FBALC_03992.
3.4. EGID Analysis
Five entire bacterial genomes that code six PS-LRR proteins have been determined [47,49,50,53]. The EGID program [39] utilizing six computational tools predicts that Synechococcus sp. CYA_1022 was transferred from another organism. That is, this protein is a paralog, not an ortholog.
3.5. Database Similarity Search of Domains including AhpC-TSA and IgA Peptidase M64 within Four Bacterial PS-LRR Proteins
F. bacterium BACCOP_00862 consists of three tandem domains-PS-LRRs, AhpC-TSA[62], and IgA peptidase M64 [63]-as noted (Figure 1). A database similarity search using the amino acid sequences of the PSLRRs as the probe identified thirty plant PS-LRR proteins with significant similarity (E-values < 10−15). In contrast, a database similarity search using the AhpCTSA and IgA peptidase M64 domains as probes detects no eukaryotic protein; the peptidase M64 domain identifies seventeen bacterial proteins with significant similarity (E-value < 10−5). Moreover, IgA peptidase M64 domain in BACCOP_03537, immunoglobulin domain in Fjoh_0602, Calx-b and He_PIG domains in CwatDRAFT_2187 detect only bacterial proteins but no eukaryotic (plant) proteins with significant similarity.
4. DISCUSSION
4.1. HGT of Tandem PS-LRRs between Plants and Bacteria
The results from the present analyses of bacterial PSLRR domains and PS-LRR proteins are summarized:
(1) PS-LRR proteins are widely distributed in most (or all) plants; they are found in only a few bacterial species. There are no PS-LRR proteins in archaea.
(2) Amino acid sequence analyses of all bacterial, candidate PS-LRRs detected here reveal that these PS-LRRs clearly belong to a “plant specific” LRR class (Figure 2). These bacterial LRRs are significantly more similar to PS-LRRs than to the other seven classes and to other LRRs of bacteria.
(3) Nucleotide similarity searches using the sequences coding the bacterial PS-LRRs reveal that the LRRs in the nine bacterial proteins are quite similar to the PS-LRRs in the 83 eukaryotic proteins with highly significant similarity (E-values < 10−10). The PS-LRRs in some bacterial proteins are more similar to those in eukaryotic proteins than to those in other bacterial proteins.
(4) The EGID program predicts that Synechococcus sp. CYA_1022 came from another organism.
(5) Out of thirty-one cyanobacterial species of which the entire genomes have been determined, only two (Synechococcus and C. watsonii) contain PS-LRR proteins.
(6) Database searches of AhpC-TSA, IgA peptidase M64, the immunoglobulin domain, the Calx-b domain, and the He_PIG domain within four bacterial PS-LRR proteins show no similarity with any eukaryotic (plant) protein, in contrast to the similarities of their respective PS-LRRs.
(7) Most of these bacteria that have PS-LRR proteins are found in aqueous environments (Table 1). Beggiatoa lives on the rice rhizosphere [66,67]: and Falavobacterium ginsengiterrae sp.nov, which shows the highest similarity with F. johnsoniae UW101 (97.1%) containing two PS-LRR proteins (Table 1), is a commensal bacteria of ginseng plant [68].
The results (1) - (3) provide evidence for the occurrence of HGT between plant and bacterium. Taken together with the results (1) - (3), the results (4) - (5) give the most parsimonious scenario that the bacterium involved in the HGT is a cyanobacterial species—Synechococcus sp. The result (6) also supports the HGT between plant and bacterium, because a commensal relationship
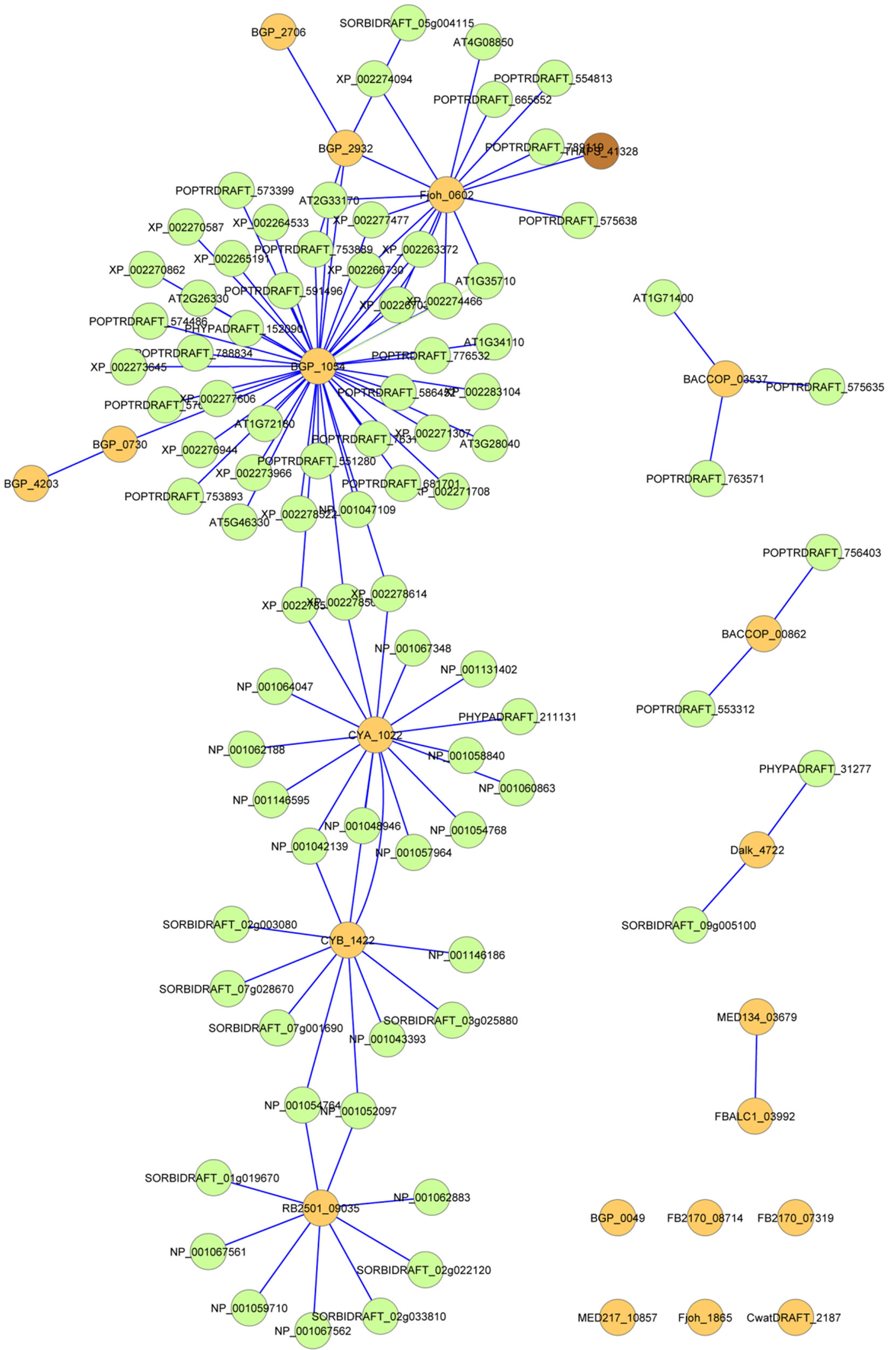
Figure 4. A similarity network of nucleotide sequence of PS-LRRs in the 20 bacterial PS-LRR proteins and the 83 eukaryotic PS-LRR proteins. Straight lines were drawn among the PS-LRR proteins in which nucleotide sequences coding the LRRs have highly significant similarity with E-values < 10−10 . Orange color is bacteria, green is plants and brown is the diatom, Thalassiosira pseudonana.
between Beggiatoa and rice, the result (7), should facilitate the HGT.
If PS-LRRs diverged from a single ancestral gene, there might be subsequent loss of that gene in many bacteria and in many eukaryotes. The origin of this single ancestral gene would have occurred prior to the divergence of archae, ~3,000,000,000 ybp. The conserved residues of PS-LRR are characterized by the degeneracy of the genetic code; Leu, which occurs at positions 1, 4, 6, 11, and 22, uses six codons; Pro, position 16, and Gly, positions 13 and 20, have four codons. Many synonymous substitutions in the PS-LRRs would have occurred over long periods. Thus, the result (3) indicates that this divergent hypothesis is improbable. If this divergence is not true, the alternative is multiple ancestors and convergent evolution; the ancestors would be a single ancestral gene for the bacterial PS-LRRs and a single ancestral gene for the eukaryotic PS-LRRs. This hypothesis conflicts the result (3).
The possibility of HGT of GALA-LRR from plant to bacteria has been reported [14]; the transferred gene was proposed to encode the F-box domain, a motif of about 50 amino acids that mediates protein—protein interactions [69,70]. This possibility is based on both structural modeling of GALA-LRRs and phylogenetic trees (using amino acid sequences of F-box domain plus the downstream F-box adjacent region or 2 - 3 LRRs) with low average branch support values [14]. The HGTs of “TpLRR” and some other LRRs between eukaryotes and bacteria have been also inferred [27,28].
4.2. Direction of HGT of Tandem PS-LRRs
There are two hypotheses for the direction of the HGT between plants and bacteria. One is that there was HGT of (part of) a PS-LRR protein from a bacterium to an ancestor of all plants. This PS-LRR protein was retained in most plants but lost in most bacteria. The HGT would have been a very ancient event, ~1,500,000,000 ybp, prior to the divergence of plant species. It is impossible to understand the result (3), as explained already. It is well recognized that the cyanobacteria formed the plastid endosymbiont and then many genes from the original endosymbiont were transferred to the host nucleus [71, 72]. The PS-LRR might maintain its structure over extended periods after the formation of this endosymbiont, since PS-LRR is an important functional element. If it is true, the PS-LRR protein would be contained in all or almost cyanobacterial species. However, this conflicts with the result (5), as well as the result (3).
The second hypothesis is that there was a more recent HGT of (part of) a PS-LRR protein from plant to bacterium. This hypothesis is consistent with all of the results (1) - (7). The result (6) suggests that the recipient bacterium in the plant-to-bacterium HGT hypothesis might be C. watsonii, F. johnsoniae or B. coprocola.
The genes encoding plant PS-LRR proteins are frequently free of introns within their respective tandem PSLRR domains (data not shown). Gene fragment acquisition appears to be common in microbial genomes [73]. In eukaryote to bacterium transfers, capturing a gene piece rather than a complete open reading frame (ORF) is more likely due to the possible presence of introns in the ORF [28]. Thus, the present results provide strong evidence for HGT of genes/gene fragments encoding PSLRR domains from plants to bacteria. The plant-to-bacterium HGT event(s) seems more probable, since the separation of the soma and germ line in multicellular organisms is generally expected to result in a very low frequency of gene transfer events into germ line cells [29, 74].
4.3. HGT of Tandem PS-LRRs between Bacteria
Bacterial tandem PS-LRRs are present in the eleven bacterial species (Table 1). The present analyses give one other result.
(8) Nucleotide similarity searches using the sequence coding the bacterial PS-LRRs reveal that bacterial PSLRRs are quite similar among the eleven bacterial species in the three different phyla.
The result (8) indicates that HGT events occurred among the eleven bacterial species, of the three phyla, as well as the results (3) and (4) (Figure 4). There are many other examples of HGTs between bacteria [75,76].
Four bacterial proteins including F. bacterium BACCOP_00862 have domains such as AhpC-TSA and IgA peptidase M64 (Figure 1) that may result from fusion events between genes/gene fragments encoding PSLRRs and other genes unique to bacteria after the PSLRR HGT event.
4.4. Evolutionary Scheme of HGTs of Tandem PS-LRRs
The most parsimonious scenario, based on the results (1) - (8), is that at least one HGT event of PS-LRR genes occurred from plant to bacteria, and subsequent HGTs occurred among the eleven bacterial species (Figure 5). A series of HGTs might have occurred recently and rapidly. We emphasize that the HGT events occurred at least two times—at least once from plant to bacterium and at least once between bacteria.
All PS-LRR proteins identified here consist of tandem LRRs of which the repeat number ranges from 2 to 43 (Figure 1 and Appendix 3). The phylogenetic inference on the whole proteins for large numbers of divergent taxa appears highly problematic [14]. We, therefore, did not perform the phylogenetic analysis. Future studies should
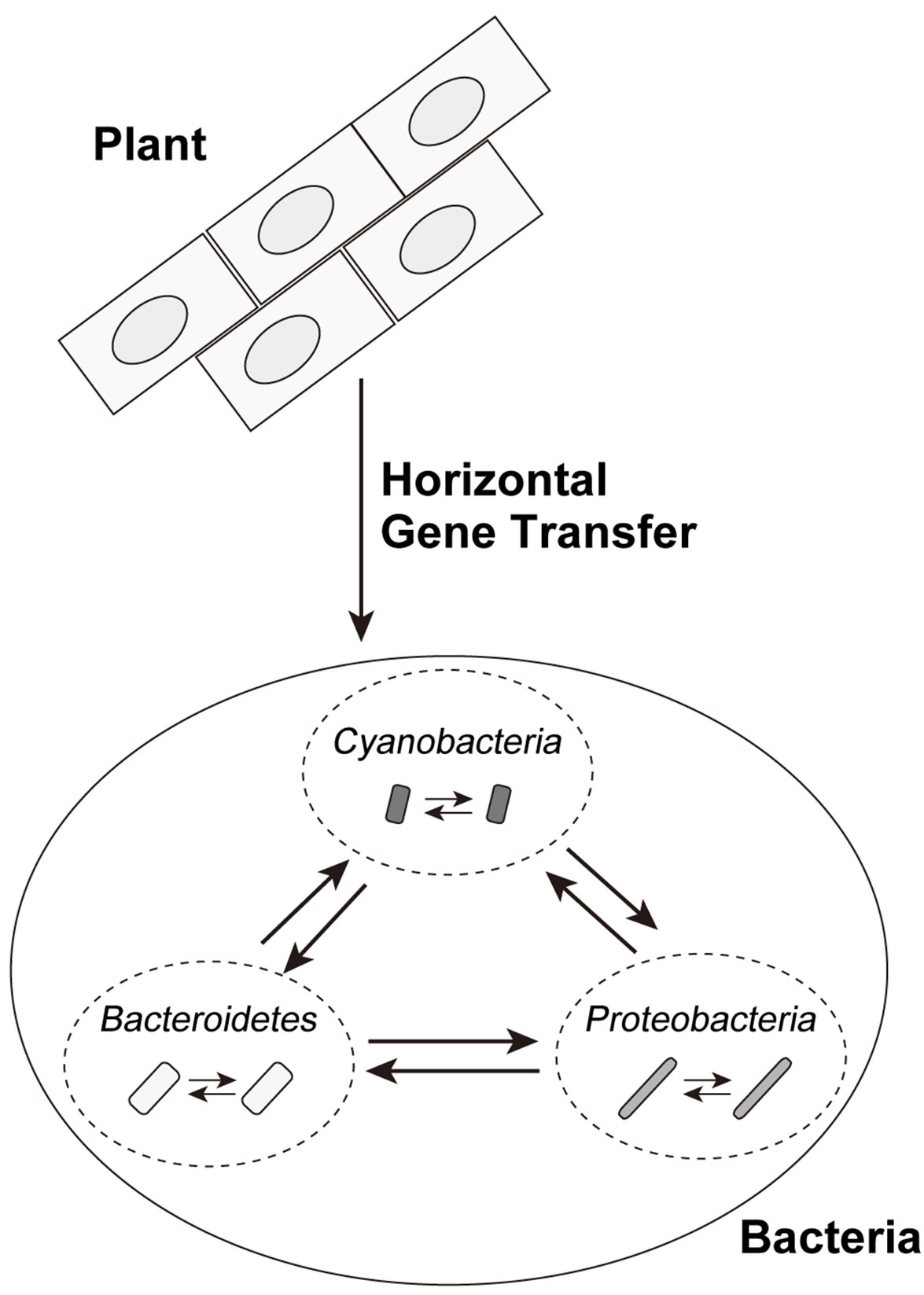
Figure 5. Possible horizontal gene transfers of bacterial PS-LRR domains. The solid line arrows show directions of possible HGT events.
resolve this problem.
5. CONCLUSION
In conclusion, analyses of the distribution of organisms having PS-LRR proteins, of similarity searches of both amino acids sequences and nucleotides sequences, as well as results of the program EGID, indicate that at least one HGT event of genes/gene fragments encoding PS-LRR domains between bacteria and plants, and HGT among the eleven bacterial species, or the three phyla, as opposed to descent from a common ancestor. There is the possibility of the occurrence of one HGT event from plant to bacteria. A series of HGTs among bacteria might have occurred more recently.
6. ACKNOWLEDGEMENTS
This study was supported by a grant from a Grant-in-Aid for Scientific Research (C) No. 23500368 from the Japan Society for the Promotion of Science (to N. M).
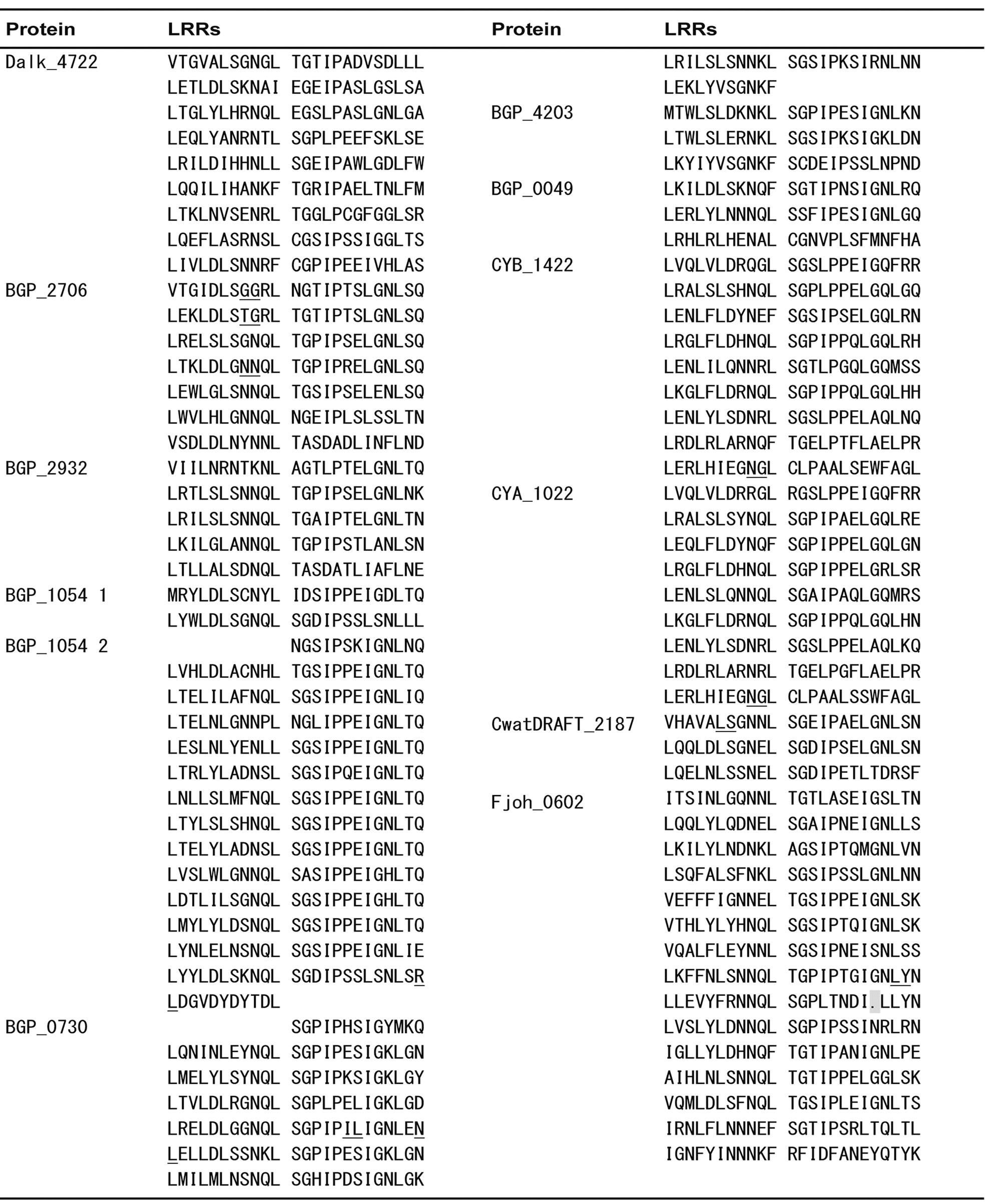
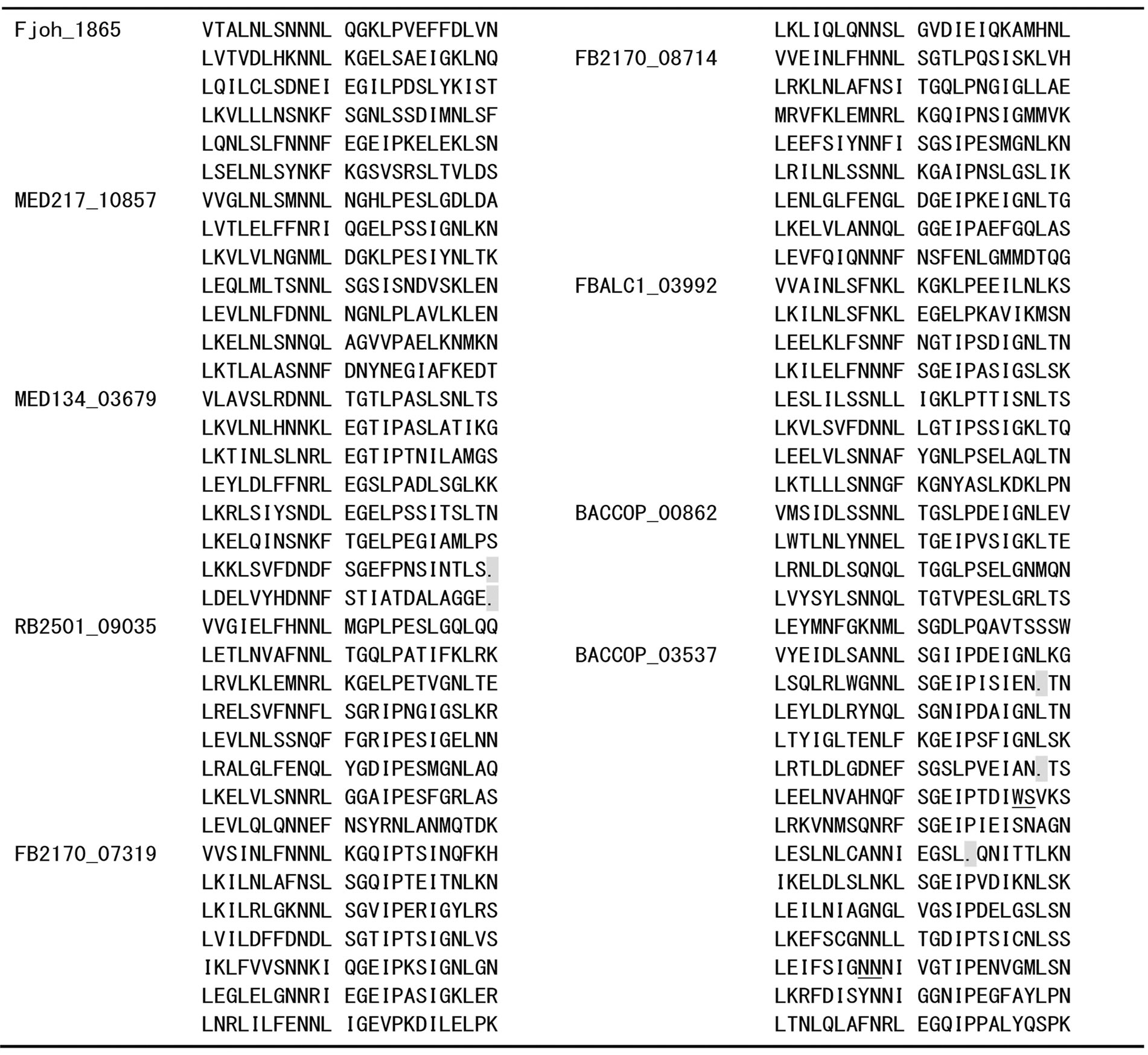
Appendix 1 . Amino acid sequence alignment of PS-LRRs within the twenty bacterial proteins.
Residues used for the PS-LRRs are 83 - 298 for Dalk_4722 with 629 residues (res.), 80 - 278 for BGP_2706 with 1,094 res., 77 - 196 for BGP_2932 with 615 res., 1 - 48 and 174 - 510 for BGP_1054 with 1,308 res., 7 - 203 for BGP_0730 with 254 res., 1 - 72 for BGP_4203 with 102 res., 7 - 78 for BGP_0049 with 362 res., 62 - 278 for CYB_1422 with 295 res., 68 - 284 for CYA_1022 with 296 res., 358 - 431 for CwatDRAFT_2187 with 927 res., 150 - 509 for Fjoh_0602 with 2,491 res., 65 - 208 for Fjoh_1865 with 237 res., 64 - 231 for MED217_10857 with 241 res., 64 - 253 for MED134_03679 with 253 res., 75 - 266 for RB2501_09035 with 302 res., 64 - 255 for FB2170_07319 with 294 res., 69 - 260 for FB2170_08714 with 295 res., 53 - 244 for FBALC1_03992 with 271 res., 51 - 170 for BACCOP_00862 with 672 res., and 348 - 697 for BACCOP_03537 with 1,049 res.. Sequences with insertions between underlined pairs of residues were removed for convenience. Deletions are indicated by dots.
Appendix 2.Multiple sequence alignments of the putative N-cap/LRRNT regions of tandem PS-LRRs in bacterial proteins. (a) Residues used for the N-cap regions are 25-82 for Dalk_4722, 24-79 for BGP_2706, 21-76 for BGP_2932, 104-173 for BGP_1054, 284-357 for CwatDRAFT_2187, 97-149 for Fjoh_0602, 20-64 for Fjoh_1865, 19-63 for MED217_10857, 19-63 for MED134_03679, 30-74 for RB2501_09035, 19-63 for FB2170_07319, 24-68 for FB2170_08714, 1-52 for FBALC1_03992, 1-50 for BACCOP_00862, and 301-347 for BACCOP_03537; (b) Residues used for the N-cap regions are 1-61 for CYB_1422, and 1-67 for CYA_1022.
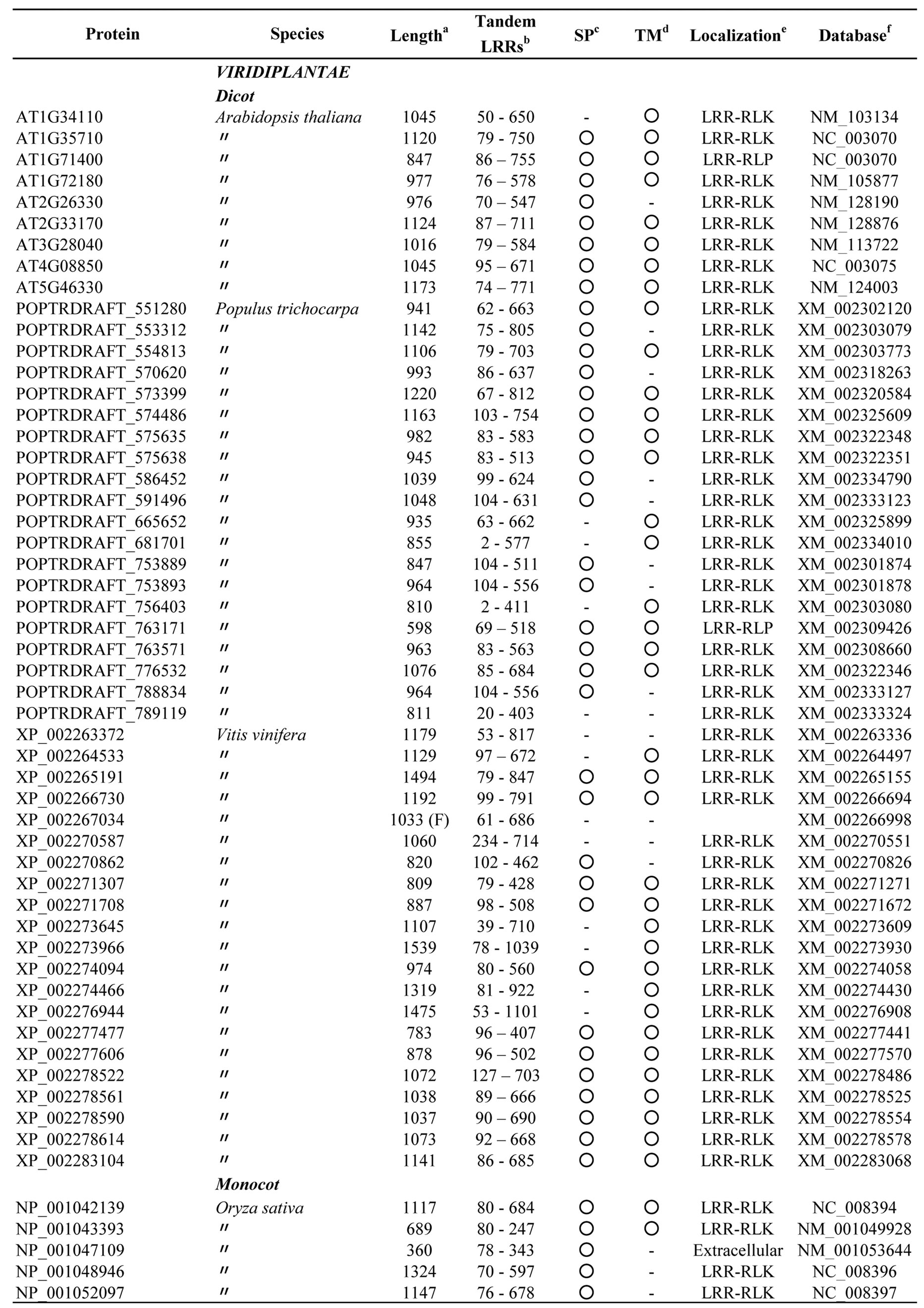
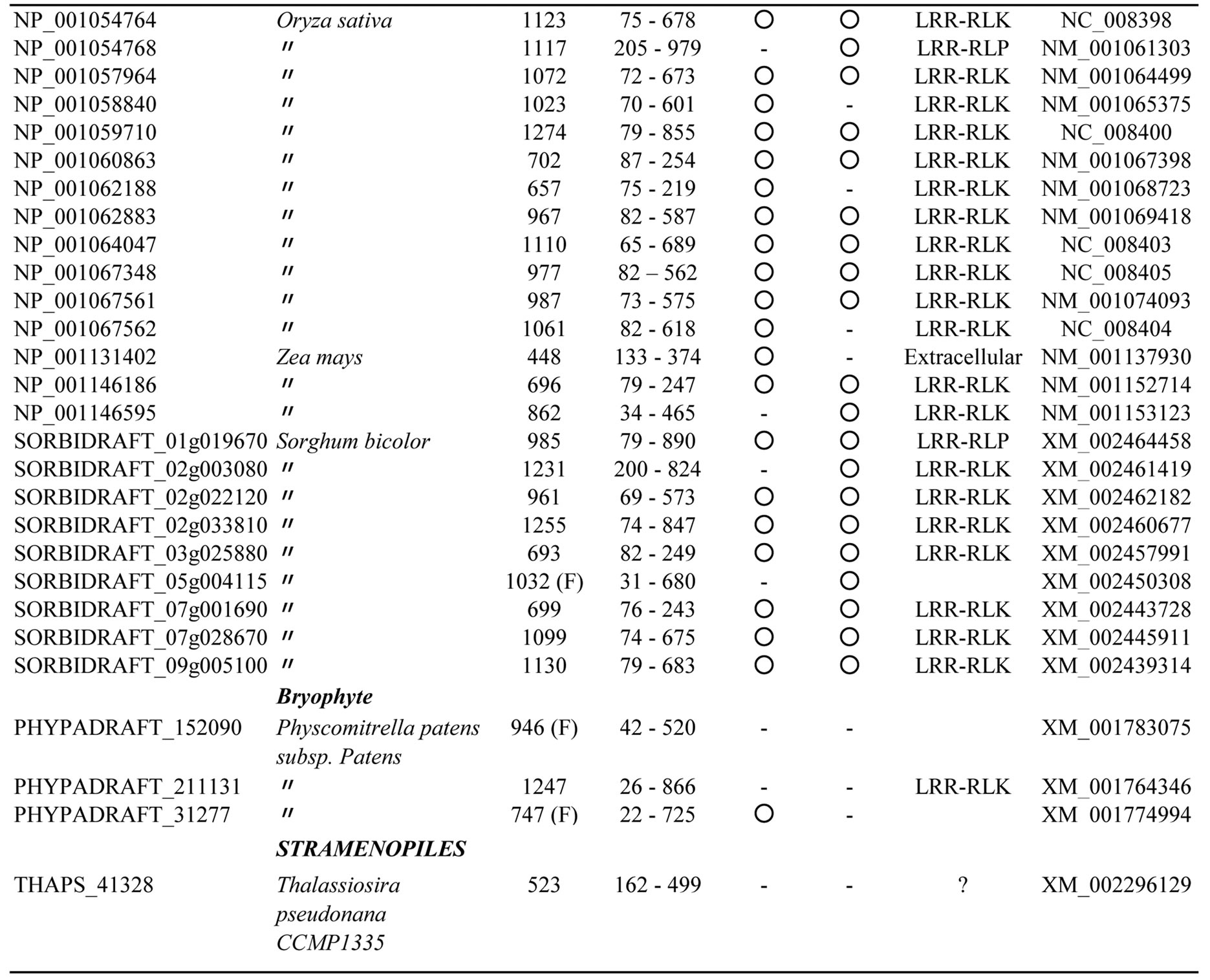
Appendix 3.Characterization of eukaryotic proteins having bacterial “PS-LRR” domain.
a: The length of complete amino acid sequences of proteins; b: The residue number in the tandem LRRs; c: Signal peptide. The occurrence (○) and non-occurrence (-) of signal peptide sequence; d: Transmembrane. The occurrence (○) and non-occurrence (-) of a transmembane region; e: The localization site in the cell, LRR-RLK or LRR-RLP; f: Protein accession number or identification number in EMBL or NCBI.