Agricultural Sciences
Vol.09 No.03(2018), Article ID:83280,23 pages
10.4236/as.2018.93025
Compositional Shifts in Ammonia-Oxidizing Microorganism Communities of Eight Geographically Different Paddy Soils
―Biogeographical Distribution of Ammonia-Oxidizing Microorganisms
Lu Lu1*, Huilin Li2#, Yan He2, Jing Zhang1, Juan Xiao1, Chao Peng1
1College of Environmental Science and Engineering, China West Normal University, Nanchong, China
2Key Laboratory of Southwest China Wildlife Resources Conservation College of Life Sciences, China West Normal University, Nanchong, China

Copyright © 2018 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: February 22, 2018; Accepted: March 23, 2018; Published: March 26, 2018
ABSTRACT
Soil nitrification is mediated by ammonia-oxidizing archaea (AOA) and bacteria (AOB), which occupy different specialized ecological niches. However, little is known about the diversification of AOA and AOB communities in a large geographical scale. Here, eight paddy soils collected from different geographic regions in China were selected to investigate the spatial distribution of AOA and AOB, and their potential nitrification activity (PNA). The result showed that the abundance of AOA was predominant over AOB, indicating that the rice fields favor the growth of AOA. PNA highly varied from 0.43 to 3.57 μg NOX-N∙g∙dry∙soil∙h−1, and was positively related with soil NH3 content, the abundance of AOA community, and negatively related with the diversity of AOB community (P < 0.01), which indicating that AOA might be the more dominant ammonia oxidizers in the collected paddy soils. Denaturing gradient gel electrophoresis fingerprints of amoA genes revealed remarkable differences in the compositions of AOA and AOB community. Phylogenetic analyses of amoA genes showed that Nitrosospiracluster-3-like and Nitrosomonas cluster 7-like AOB extensively dominated the AOB communities, and 54d9-like AOA within the soil group 1.1b predominated in AOA communities in paddy soils. Redundancy analysis suggested that the spatial variations of AOA community structure were influenced by soil TN content (P < 0.01), while no significant correlation between AOB community structure and soil properties was found. Findings highlight that ammonia oxidizers exhibit spatial variations in complex paddy fields due to the joint influence of soil variables associated with N availability.
Keywords:
Paddy Soil, Ammonia-Oxidizing Microorganism, Nitrification Activity, Large Geographical Scale, Diversification
1. Introduction
Microbial ammonia oxidation is the first and rate-limiting step of nitrification, which is the only oxidative process that connects the oxidized and reduced state of inorganic nitrogen to maintain the global nitrogen cycling [1] . The discovery of ammonia-oxidizing archaea (AOA) has fundamentally expanded our perceptions of nitrification, which has long been thought to be restricted to ammonia-oxidizing bacteria (AOB), which are phylogenetically belonging to Proteobacteria [2] . Although recent evidence demonstrated Nitrospia bacteria also has the complete nitrification system [3] , the ubiquity of AOA and AOB across a wide variety of environment and habitats implies that metabolically distinct ammonia oxidation microorganisms play critical roles in nitrification and may be adapted to live under physicochemical contrasting niches [4] . Numerous researchers have recognized that the functionally relevant diversity of AOA has greatly outnumbered their bacteria counterparts [5] [6] . AOA has been proved to be the dominant group in nitrification in some soils [7] [8] , and in some acidic soils which only AOA are detectable [6] [9] , while the converse has also been demonstrated in agricultural soils [10] and N-rich soils [11] . These inconsistent results raise continuous debate about the contribution of AOA and AOB to nitrification [12] . So far, the environmental factors that drive diversification of ammonia-oxidizing microorganism remain largely unexplored, especially anthropogenically disturbed ecosystems. The large-scale geographic analysis of AOA and AOB population may provide intriguing hints regarding the niche differentiation of ammonia oxidizer communities and their contribution in nitrification.
Rice fields occupy a large portion of global cropland and nearly account for 26% of China’s total croplands [13] . Nitrification in paddy soils contributes a lot to the nitrogen cycling in the terrestrial ecosystem. The paddy soils are subjected to temporal or permanent flooding during the growth of rice, which makes them model systems to investigate the divergence of ammonia oxidizers communities in microaerobic niches of the surface soil. Cultured-based assays indicated that AOA can be enriched under such circumstances [14] . AOA which exhibit the capacity of detoxification of reactive oxygen species (ROS) may support them to be active in the microaerobic environment [15] . amoA-based quantification showed that AOA was more abundant in the paddy soils [16] [17] . In consideration of those data, it seems that AOA might be more active to the paddy soils rather than AOB.
In addition to oxygen fluctuations, much is now known about the multiple environmental factors which were reported to regulate the community structures of AOA and AOB, including ammonia substrate availability [18] , pH [19] , temperature [20] , moisture [21] , and soil organic carbon [22] . Among these factors, many studies have stated that pH might be the most dominant variable to govern the niche differentiation of AOA and AOB [12] . Acidophilic AOA isolate Nitrosotalea devanaterra showed extraordinary affinity to ammonia substrate and grow at an extremely low ammonia concentration of 0.18 nM [23] , while the activity of AOB was found to decrease with pH and could only grow on ammonia at μM level [24] . It has also been demonstrated that long-term crop rotation, fertilization, and tillage can impact the activity and structure of ammonia-oxidizing communities [25] . Nevertheless, little is known about the extent of paddy soil heterogeneity could have shaped the differentiation of AOA and AOB communities.
Large geographical scale experiment may provide comprehensive insights into how different types of soils with various chemical properties lead to the diversification of ammonia-oxidizing communities. The present study was conducted on eight paddy soils which geographically distribute in different temperature zone, from cold temperate zone to subtropical zone, and also have a rice cultivation history of >15 years with a wide pH range of 5.37 - 7.99. We employed quantitative real-time PCR and DGGE combined with clone library approaches based on the amoA gene to explore the correlation between variable soil properties and activity and community structure of ammonia-oxidizing microorganisms.
2. Material and Methods
2.1. Site Description and Soil Sampling
A total of eight paddy soil samples representing eight parent material types from different provinces were collected from northern China to southern China during September, 2016, to exclude seasonal variation (Figure 1 and Table S1). These paddy soil samples were obtained from Heilongjiang (HLJ), Beijing (BJ), Shanxi (SX), Henan (HeN), Anhui (AH), Jiangxi (JX), Chongqing (CQ) and Hunan (HuN), respectively. The types of soils were indicated in Figure 1. The paddy field all had a history of rice cultivation for more than 15 years. At each site, triplicate surface soil samples (depth, 0 to 15 cm) were randomly obtained by using a sterile hand trowel. Collected samples were placed in sterile plastic bags, sealed, and transported to the laboratory on ice. Plant residues and other materials, such as stones and obvious macrofauna, were removed before each replicate from each field was homogenized through a 2 mm mesh. Aliquots (5 g wet weight) of soil from each site were stored at −80˚C prior to subsequent DNA extraction and molecular analysis. The subsamples were stored at 4˚C for chemical determination and potential nitrification activity (PNA). PNA was processed immediately after arrival.
Figure 1. Geographic map of sampling sites.
2.2. Soil Physiochemical Properties Analysis
Chemical characteristics including pH, soil water content (SWC), -N, -N, soil organic matter (SOM) and total nitrogen (TN) were determined in this study. The soil pH was determined using a Mettler Toledo 320-S pH meter (Mettler-Toledo Instruments Co.Ltd., Shanghai, China) with a water-to-soil ratio of 2.5. Water content was determined gravimetrically by drying the soil at 105˚C for 6 h. -N and -N were extracted from soil samples with 2 M KCl, and the contents were determined using a Skalar San Plus segmented flow analyzer (Skalar Inc., Breda, The Netherlands). SOM was detected using a total carbon analyzer (TOC-V CPH, Shimadzu, Japan). TN was determined by the Kjeldahl digestion method. All analyses were performed in triplicate for each soil sample. These parameters were then calculated based on oven-dried soil weight.
2.3. Nitrification Potential Measurements
The PNA is the maximum capacity of nitrifying microorganisms to transform -N to -N in a soil population. Changes in nitrification potential provide quantitative information on how nitrifying communities behave in various environmental conditions and reflect the in situ nitrification rates. Nitrification potential was determined according to the shaken-slurry method [26] . PNAs were measured with fresh soils from each site by mixing each of triplicate. 15 g (wet weight) of soil from each sample was mixed with 100 ml of 1.5 mM ammonium sulfate. After incubation for 0, 2, 6, 12 and 24 h at 25˚C, 10 ml slurry samples were centrifuged and the supernatant was filtered through a 0.45 μm pore size membrane and stored at −20˚C until analysis. -N content in the supernatant was immediately analyzed. -N concentration increased linearly, and nitrification potential (μg -N∙g−1∙h−1) was calculated from the rate of increase in -N concentration over time in the slurry using linear regression.
2.4. Soil Genomic DNA Extraction
The total DNA in soil samples was extracted and purified from 0.5 g (dry weight) of soil using a FastDNA spin kit for soil (MP Biomedicals, Cleveland, OH, USA), according to the manufacturer’s instructions. The quantity and quality of DNA extracts were assayed using a NanoDrop ND-1000 UV-visible light spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The soil DNA was stored at −20˚C for further analysis.
2.5. Real-Time qPCR of amoA and 16S rRNA Genes
Real-time quantitative PCR (qPCR) was performed in triplicate on a CFX96 Optical RealTime detection system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) to determine the copy number of amoA and 16S rRNA genes in soil DNA extracts. The number of amoA gene copies per g of soil (dry weight) was measured using primers amoA-1F and amoA-2R [27] targeting amoA gene of AOB, and primers amoA-AF and amoA-AR [28] targeting amoA gene of AOA. A universal 16S rRNA gene qPCR was conducted to assess the total copy number of bacterial communities by using the primer 515/907. The primers and PCR/qPCR thermal conditions were detailed in Table S2 in the supplemental material.
The real-time quantitative PCR standard was generated using plasmid DNA from one representative clone containing bacterial or archaeal amoA, and 16S rRNA gene. A dilution series of standard template over eight orders of magnitude per assay was used. In addition, a series dilution of soil DNA extract was also used to assess whether a PCR inhibition by humic substance co-extraction occurred in the paddy soils. The amoA gene copies decreased proportionally with the diluted soil DNA template concentrations. A 10-fold diluted soil DNA was used for subsequent analysis. qPCR was performed in a reaction mixture containing 1 × SYBR® Premix Ex Taq™ (TaKaRa Biotech, Dalian, China), 0.5 mM of each primer, and approximately 1.0 - 5.0 ng DNA template. Amplification efficiencies ranged from 91% to 103%, with R2 values of 0.993 to 0.999. Melting curve analysis and standard agarose gel electrophoresis were always performed at the end of a PCR run to verify the amplification specificity.
2.6. DGGE Analysis of amoA Gene
AOB and AOA composition were analyzed by a DGGE of amoA genes using the D-Code system (Bio-Rad Laboratories, Hercules, CA), as described previously [6] . PCR reaction was carried out in a thermal cycler (Bio-Rad Laboratories, Hercules, CA). The thermal condition was listed in Table S2. The PCR products were checked using 1.2% agarose gel to ascertain the specificity of bacterial amoA and archaeal amoA gene amplification with a size of ~489 bp and ~629 bp, respectively. The 150 to 200 ng PCR amplicons of bacterial amoA were separated using 8% (wt/vol) polyacrylamide [acrylamide-bisacrylamide (37.5:1)] gels with a denaturing gradient of 30% - 60% (100% denaturant contains 7 M urea and 40% formamide), and the 150 to 200 ng PCR amplicons of archaeal amoA were separated using 6% gels with a denaturing gradient of 30% - 50%. A 1 mm thick gel was poured from bottom to top using a gradient former and peristaltic pump at a speed of 4.5 ml∙min−1. A 5.0 ml stacking gel containing no denaturants was subsequently added on top before polymerization to insert a comb to make 24 wells. An electrophoresis separation of bacterial amoA and archaeal amoA gene amplicons was carried out using a 0.5 × Tris-acetate-EDTA buffer at 80 V for 17 h at 60˚C. The gels were stained with 1:10000 (v/v) SYBR Green I for 30 min, and then scanned with a Molecular Imager FX using Quantity One software package (Bio-Rad Laboratories, Hercules, CA, USA). Dominant bands in the DGGE fingerprints were excised and reamplified. The reamplified PCR products of the DGGE bands were cloned using pEASY-T3 vector (TransGenBiotech, Beijing, China) according to the instructions of the manufacturer. The clones containing the exact bacterial amoA and archaeal amoA gene insert from each DGGE band were sequenced (Invitrogen, Shanghai, China). DNAMAN software was used to check the clone sequences manually for subsequent analyses. Phylogenetic analysis was performed using the Molecular Evolutionary Genetics Analysis (MEGA 7.0) software package. The sequences of basic tree were from known AOB and AOA cultures, and fosmid clones of the amoA genes.
2.7. Statistical Analysis
The concentration of -N, , SOM, TN, PNA and amoA gene copy numbers were compared through multiple sample comparisons using one-way ANOVA analysis followed by Student-Newman-Keulstest to check for quantitative variance between different samples. The two-way ANOVA analysis was performed to examine the effect of different soil characteristics and soil types on PNA. Pairwise comparisons between treatments were determined with two-tailed Student’s t-tests. All analyses were conducted using SPSS version 20.0 (IBM, Armonk, NY, USA). Pearson’s correlation analyses were performed to assess the relationships among PNA, soil properties and the abundances of AOA and AOB. Redundancy analysis (RDA) with the Monte Carlo permutation’s test (499 permutations) was carried out to determine if the AOA and AOB community structures were correlated with PNA and soil properties using Canoco (version 4.5).
2.8. Accession Numbers of Nucleotide Sequences
The nucleotide sequences were deposited at the GenBank with Accession Numbers MG000601-MG000613 and MG000592-MG000600 for the bacterial amoA and archaeal amoA genes in this study, respectively.
3. Results
3.1. Soil Properties and Nitrification Activity
Soil properties including SOM, TN, inorganic nitrogen concentration, soil water content, pH were listed in Table 1. The soil samples were collected from different latitude from 21 m to 931 m. Soil samples from BJ, SX, HeN, AH, and CQ had similar pH values from 7.72 to 7.99, which was classified as alkaline soil. Acidic soil samples from HLJ, JX, and HuN had relatively lower pH values of 5.37, 5.38 and 6.10, respectively. The water content of the soil samples ranged from 10.82% to 29.37%. There was significantly difference in SOM and TN content of these soil samples from different sampling sites (P < 0.05), ranging from 21.34 to 49.11 g∙kg−1 and 0.68 to 1.23 g∙kg−1, respectively, with the highest content in HLJ sample. SX sample had the highest -N and -N content, while CQ sample had the lowest -N content and JX had the lowest -N content. Soil NH3 concentrations were estimated to be varied 0.41 to 553.18 μM. The NH3 content was lowest in the acidic soil sample JX and was highest in the alkaline soil sample AH. This is consistent with the principle that NH3 is ionized exponentially to with decreasing pH [29] . The PNA ranged from 0.43 to 3.57 μg -N∙g−1∙h−1 (Table 1) across all paddy soils. Sample from AH was found to have the high PNA. As revealed by Pearson correlation analysis, PNA had significant relationship with soil NH3 concentration (r = 0.82, P < 0.01) in Table 2.
3.2. Abundance of Bacterial and Archaeal 16S rRNA Gene and amoA Genes
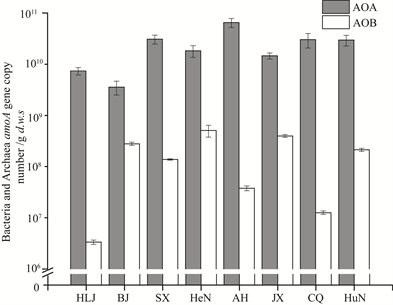
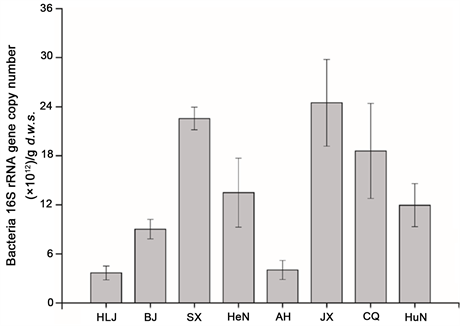
Bacterial 16S rRNA and amoA gene were quantified by qPCR to investigate the variation in population sizes of microbial and nitrifying communities (Figure 1S and Figure 2). As shown in Figure 2, the copy numbers of archaeal and bacterial amoA genes ranged from 3.58 × 109 to 6.54 × 1010, and from 3.35 × 106 to 5.09 × 108 copies∙g−1 d.w.s in these paddy soils, respectively. AOA abundance was
Table 1. Sampling site and soil characteristicsa.
aMeans of three replicates per treatment are presented with standard deviation; bSOM and TN denote soil organic matte and total nitrogen, respectively. cpH was measured with water-soil ratio of 2.5. dSoil NH3 concentration was estimated on the basis of soil water content, ammonium concentration, pH and adjusted for temperature (pKa of 9.245 at 25˚C); ePNA, potential nitrification activity.
Table 2. Pearson correlation of chemical properties, PNA, abundance or ratio of bacteria, AOB and AOA.
*Correlation is significant at the 0.05 level; **Correlation is significant at the 0.05 level.
Figure 2. The abundances of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) amoA genes (represented by black and white bars, respectively). The error bars represent the standard error of the means of the triplicates. All other designations are the same as those in Table 1.
generally higher that AOB abundance in paddy soils. The ratio of archaeal to bacterial amoA gene copy numbers was varied from 12.8 to 2400.0. The abundance of bacterial 16S rRNA genes ranged from 3.67 × 1012 to 2.45 × 1013 copies∙g−1 d.w.s in different paddy soil samples (Figure S1).
Correlation analysis was performed to reveal the influence of environmental parameters on the nitrifying communities and PNA (Table 2). There was a significant positive correlation between PNA and AOA abundance (r = 0.83, P < 0.01), whereas no relationship was observed between PNA and AOB abundance (r = −0.16, P > 0.05). The PNA was also significantly correlated with soil NH3 content (r = 0.82, P < 0.01). A significant positive correlation was found between AOB population size and SOM (r = 0.65, P < 0.05) and TN (r = 0.64, P < 0.05). In addition, the abundance bacterial 16S rRNA gene was positively correlated with soil -N content.
3.3. Diversity and Structure of Ammonia-Oxidizing Bacteria and Ammonia-Oxidizing Archaea Communities
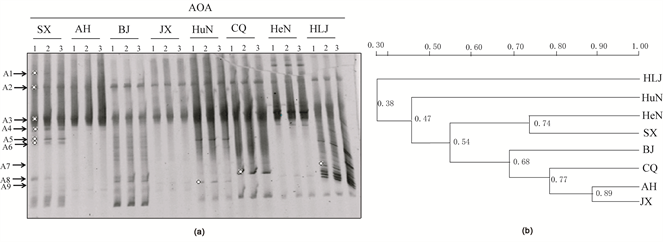
The structures of AOA and AOB communities were revealed by fingerprinting analysis of bacterial and archaeal amoA genes in all paddy soils (Figure 3(a) and Figure 4(a)). Highly reproducible DGGE fingerprints were obtained in the triplicate of each soil sample. The diversity of AOA and AOB were indicated by Shannon’s diversity index (H). richness (S) and Evenness (Eh), which were calculated by the relative abundance of various DGGE fingerprints in different soil samples (Table 3). The H and S index of AOA communities in HLJ, BJ and SX samples were clearly higher, when compared with the corresponding index of AOB, whereas the opposite trend was observed for the remaining soil samples. The highest H, S and Eh index of AOA communities was found in BJ soil, while the lowest was found in AH soil.
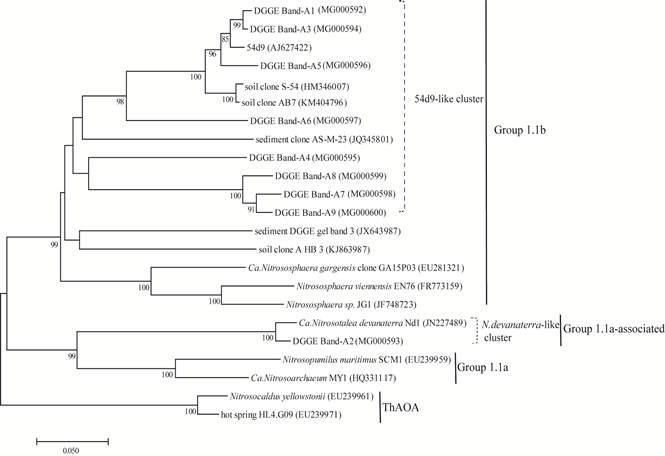
Distinct different DGGE fingerprints of archaeal amoA genes were observed among the different paddy soils. AOA communities in paddy soils were dominated by the DGGE band-A3, which were affiliated with the 54d9-like cluster of the soil group 1.1b lineage (Figure 3 and Figure 5). The majority of the DGGE bands for AOA were affiliated with the soil group 1.1b lineage except for the DGGE band-A2, which was placed within the Nitrosotaleav devanaterra-like cluster of the marine group 1.1a-associated lineage. The DGGE band-A5 and A6 showed higher intensities in the SX, HuN and HLJ samples, although they were faint in other soils. The DGGE band-A8 and A9 were observed in six soil samples except for AH and JX. The result was confirmed by cluster analysis which revealed that AH and JX samples were clearly separate from other samples with an 89% similarity (Figure 3(b)). DGGE band-A7 was only detected in HLJ.
Figure 3. DGGE fingerprints of the archaeal amoA genes in different paddy soils (a), and similarity dendrograms (UPGMA, Dice coefficient of similarity) of AOA band patterns calculated from DGGE fingerprints in different paddy soils (b). The numbers 1, 2 and 3 represent the soil triplicates of each paddy soil. The arrows indicate the DGGE bands excised for sequencing. All other designations are the same as those in Table 1.
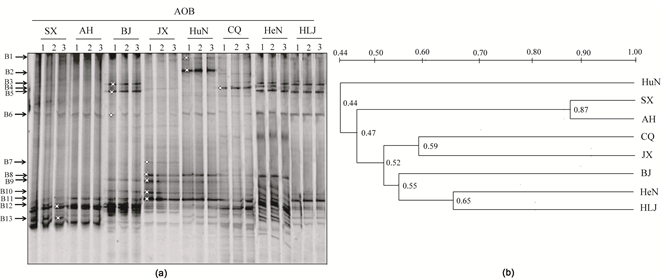
Figure 4. DGGE fingerprints of the bacterial amoA genes in different paddy soils (a), and similarity dendrograms (UPGMA, Dice coefficient of similarity) of AOB band patterns calculated from DGGE fingerprints in different paddy soils (b). The numbers 1, 2 and 3 represent the soil triplicates of each paddy soil. The arrows indicate the DGGE bands excised for sequencing. All other designations are the same as those in Table 1.
Figure 5. Phylogenetic tree showing the relationship of the archaeal amoA gene in different paddy soil. Bootstrap values higher than 80% are indicated at the branch nodes. The scale bar represents five changes per 100 nucleotide positions. GenBank accession numbers are given in parentheses.
Table 3. Shannon index richness obtained by DGGE profiles of bacterial and archaeal amoA gene in different paddy soils.
Note: Different lowercase letters in the same column mean significant difference (P < 0.05)
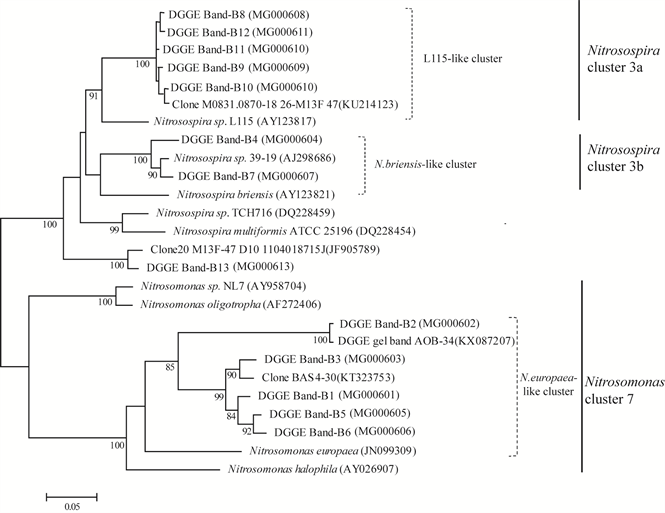
Distinct AOB communities were also observed in the paddy soils. Phylogenetic analysis showed that eight of the thirteen DGGE bands were related with the Nitrosospira cluster lineage, whereas the remaining 5 DGGE bands were placed within Nitrosomonas cluster 7 (Figure 4(a) and Figure 6). The dominant AOB DGGE bands B11, B12 and B13 with high-intensity were all clearly present in all soil samples, which were affiliated with the Nitrosospira cluster 3. Additionally, the DGGE band B3 and B5 belonging to Nitrosomonas europaea-like cluster seemed obviously higher in BJ, HeN and HLJ samples, which were also grouped into one cluster with a similarity of 55% (Figure 4 and Figure 6). The AOB communities in SX and AH samples reached 87% of the similarity. The high intensity of DGGE band-B2 affiliated with the Nitrosomonas cluster 7 was only detected in HuN sample.
3.4. Correlations of Soil Properties and PNA with Community Structures of Ammonia-Oxidizing Bacteria and Archaea
Pearson correlation analyses were performed to reveal the impact of soil properties on the diversity index of nitrifying communities and PNA (Table 4). The PNA was significantly negatively correlated with the diversity of AOB (r = 0.81, P < 0.01). The Richness E of AOA and AOB communities was positively correlated with soil SOM (AOA, r = 0.66, P < 0.05 and AOB, r = 0.64, P < 0.05) and TN (AOA, r = 0.63, P < 0.05 and AOB, r = 0.66, P < 0.05). All other diversity indexes did not show a significant correlation with the soil properties in paddy soils.
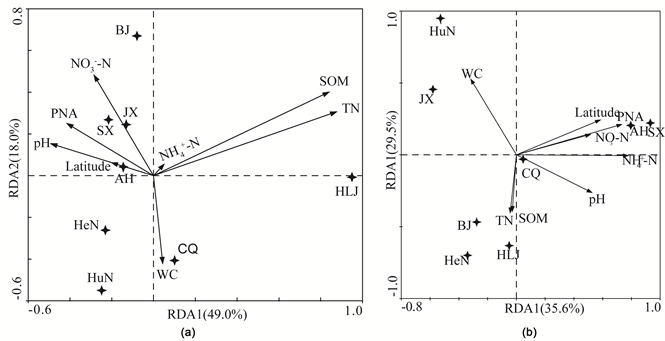
RDA was conducted to determine the correlation of soil properties with community structures of AOA and AOB (Figure 7). Partial RDAs based on Monte Carlo permutation (n = 499) only retained the significant parameters in the models. The first and second axes accounted for 49.0% and 18.0% of the total variation in AOA community structure, respectively (Figure 7(a)). Regarding
Figure 6. Phylogenetic tree showing the relationship of the bacterial amoA gene in different paddy soil. Bootstrap values higher than 80% are indicated at the branch nodes. The scale bar represents five changes per 100 nucleotide positions. GenBank accession numbers are given in parentheses.
Table 4. Pearson’s correlation coefficients of soil properties, PNA with community diversity of ammonia-oxidizing bacteria and archaea.
*Indicate significant level at p < 0.05 level. **Indicate significant level at p < 0.01 level.
Figure 7. Correlation of soil properties with community structure of AOA (a) and AOB (b) determined by redundancy analysis (RDA). The four-pointed star represents the soil sample. Arrows indicate the direction and magnitude of variables.
the AOA community, the most influential constraining variable 40% was TN (F = 4.04, P = 0.002) (Figure 7(a) and Table 5). The first and second axes accounted for 35.6% and 29.5% of the total variation in AOB community structure (Figure 7(b)). No significant correlation between AOB community structure and soil properties was found.
4. Discussion
The great demand for rice in Asia has prompted the intensified application of synthetic nitrogen chemical fertilizers to increase the rice productivity [30] . This agricultural intervention combined with flooding management is the major ecological force to drive the diversification of AOA and AOB communities. Our results exhibited the overview of phylogenetically and functionally distinct AOA and AOB communities in the 8 different paddy soils of China. This study suggests the strong adaptation of the versatile and diverse ammonia oxidizers to markedly different soil environmental conditions in the collected paddy soils.
The ratio of AOA to AOB in this study ranged from 12.8 to 2400.0. It appears that the paddy soils with agricultural interventions, such as frequent flooding and fertilizations, favored the growth of AOA communities. Our results agreed well with recent findings regarding ammonia oxidizer dynamics in a red paddy soil with the AOA/AOB ratio of 36.0 - 1686.0 [31] and other paddy soil in China [32] . In our study, the abundance of AOA had negative correlation with SOM and TN, which was also observed previously in surface soil [33] . This might be due to the increase of bioavailability of carbon and nitrogen that stimulate the
Table 5. Eigen values, F values and P values obtained from the partial RDAs testing the influence of the significant parameters on the AOA and AOB community compositiona.
aPartial RDA based on Monte Carlo permutation (n = 499) kept only the significant parameters in the models; bSum of all Eigen values for both partial RDA were 1.000. cF and P values were estimated using Monte Carlo permutations. dP < 0.05 indicates the significant effect.
growth of heterotrophic bacteria, and thus inhibit the chemoautotrophic ammonia oxidizers [34] . Although AOA might be capable of mixotrophic or heterotrophic growth as previously suggested [12] [35] , the ecological significance of a heterotrophic lifestyle for AOA in complex environments remains poorly understood. In contrast, positive correlations were found between SOM, TN and AOB abundance in the test soils. This nutrient-induced stimulation of AOB abundance further supports the idea that AOB could play an important role in environments with high fertilizer and nitrogen loads [36] [37] . The abundance of AOA rather than AOB was positively significantly correlated with PNA, demonstrating that AOA might be the more dominant nitrifiers in the collected paddy soils, which was also consistent with previous studies [38] .
DGGE fingerprinting analysis showed that the community structures of AOA were remarkable variable in the test soils. The dominant composition of AOA fell within 54d9-like cluster belong to soil group 1.1b (Figure 5). Pester et al. [39] also indicated that the amoA gene of soil metagenome fragment 54d9 was widely distributed, which was detected in 75% of the 146 soils covering different biomes, like forest, grasslands, deserts and agricultural soils. The existence of AOA DGGE band-A2 affiliated with N. devanaterra-like cluster indicated the AOA belonging to Group 1.1a could adapt and survive in more diverse conditions [40] . Flooding management in rice field leads to rapid depletion of oxygen beneath the soil surface. A number of studies have shown that AOA may have higher substrate affinity for oxygen than AOB [41] [42] , and be prone to adapt to life under low oxygen [43] . A marine AOA even sustains high ammonia oxidation activity at <10 μM O2 concentrations in the suboxic region [44] . RDA revealed that soil TN content significantly affected AOA community (P < 0.01) (Table 5), which is consistent with previous study [45] . Studies stated that AOA within the marine group 1.1a and 1.1a-associated lineages have higher ammonia affinity [8] [46] , whereas AOA within the soil group 1.1b lineage could tolerate high ammonium concentrations [47] . These findings indicate that the N available in the paddy soil could be a key parameter to shape the diversification of AOA community.
DGGE fingerprinting analysis combined with phylogenetic analyses demonstrated that AOB affiliated with the Nitrosopira cluster-3a and 3b dominated the communities of AOB in the test soils, which was consistent with the previous studies in paddy soils [48] [49] . It was previously proved that Nitrosospira cluster 3 - like AOB performed most bacterial ammonia oxidation in some Chinese paddy soils [49] . AOB belonging to Nitrosospira cluster 3 often outcompete other Nitrosospira species under high ammonia conditions [50] . The highly enrichment of Nitrosospira cluster 3 in these paddy soil could be due to the anthropogenic management such as irrigation and fertilization. Moreover, AOB affiliated with Nitrosomonas europaea (cluster 7) was also observed in some of the test paddy soils except AH and SX samples. The detection of Nitrosomonas-like AOB in paddy soils was in agreement with previous studies [48] [51] , whereas in some studies only Nitrosospira-like AOB was detected [45] [52] . The rice paddy ecosystems with regular flooding, which could lead to strong fluctuations in available oxygen. Some members of Nitrosomonas has high affinity for oxygen, supporting them to be dominant in the micor-oxic environment [53] , and the extensive present of Nitrosomonas-like AOB in the test paddy soils. Moreover, the different pattern of AOB community structures in these paddy soils might be due to the different ecological niches of Nitrosomonas-like and Nitrosospira-like AOB shaped by environmental factors. It was supported by the positive correlation between soil SOM (r = 0.64, P < 0.05), TN (r = 0.66, P < 0.05) contents, and AOB community richness (Table 4). However, a higher resolution of the AOB communities and their major influence factors are required to be further studied with more powerful analysis techniques.
PNA representing the soil ammonia oxidation activity varied among different paddy soils. The positive correlation between PNA and NH3 content, the abundance of AOA community, the diversity of AOB community. This finding could be attributed to the dependence of monooxygenase on NH3, which has been thought to be the actual substrate for ammonia oxidizers [29] [54] . The positive correlation between PNA and NH3 content indicated that soil might be the limiting factor during nitrification in the test paddy soils. However, rice fields are generally subjected to drastic anthropogenic disturbances with fertilization and crop cultivation. The intensified fertilization was found to stimulate soil nitrification and the activity of ammonia oxidizers in neutral soil [55] and acidic soil [56] , which thus promoting the demand of and NH3 substrate. Therefore, the changes of AOA and AOB population, and these soil physicochemical variables associated with N availability together determine the PNA in different paddy soils.
5. Conclusion
Taken together, our study demonstrates the differential abundance and community structure of AOA and AOB in paddy soils from northern China to southern China. The abundance of AOA was significantly predominant over AOB. A positive correlation was observed between the PNA and soil NH3 content, AOA abundance, while a negative correlation was detected between PNA and the diversity of AOB community (r = −0.81, P < 0.01). The results imply that AOA might play a more important role in controlling nitrification activity in these alkaline and acidic paddy soils. The coexistence of diverse AOA and AOB population further suggested that nitrification might be performed by different ammonia-oxidizing phylotypes combinations. This study also demonstrated that the contrasting soil physicochemical parameters of the geographically different paddy soils could synthetically determine the spatial variation of AOA and AOB communities.
Acknowledgements
This work was financially supported by the National Science Foundation of China (41606142) and the Fundamental Research Funds of China West Normal University (463140 and 412554). We thank Yan He and the local farmers for the paddy soil sample collection, and Mingli Liao, Zaijun Yang for their assistance in chemical analysis.
Cite this paper
Lu, L., Li, H.L., He, Y., Zhang, J., Xiao, J. and Peng, C. (2018) Compositional Shifts in Ammonia-Oxidizing Microorganism Communities of Eight Geographically Different Paddy Soils―Biogeographical Distribution of Ammonia-Oxidizing Microorganisms. Agricultural Sciences, 9, 351-373. https://doi.org/10.4236/as.2018.93025
References
- 1. Kowalchuk, G.A. and Stephen, J.R. (2001) Ammonia-Oxidizing Bacteria: A Model for Molecular Microbial Ecology. Annual Review of Microbiology, 55, 485-529.https://doi.org/10.1146/annurev.micro.55.1.485
- 2. Treusch, A.H., Leininger, S., Kletzin, A., Schuster, S.C., Klenk, H.P. and Schleper, C. (2005) Novel Genes for Nitrite Reductase and Amo-Related Proteins Indicate a Role of Uncultivated Mesophilic Crenarchaeota in Nitrogen Cycling. Environmental Microbiology, 7, 1985-1995. https://doi.org/10.1111/j.1462-2920.2005.00906.x
- 3. Daims, H., Lebedeva, E.V., Pjevac, P., Han, P., Herbold, C., Albertsen, M., Jehmlich, N., Palatinszky, M., Vierheilig, J. and Bulaev, A. (2015) Complete Nitrification by Nitrospira Bacteria. Nature, 528, 504-509.
- 4. Martens-Habbena, W., Berube, P.M., Urakawa, H., de la Torre, J.R. and Stahl, D.A. (2009) Ammonia Oxidation Kinetics Determine Niche Separation of Nitrifying Archaea and Bacteria. Nature, 461, 976-979. https://doi.org/10.1038/nature08465
- 5. Stahl, D.A. and de la Torre, J.R. (2012) Physiology and Diversity of Ammonia-Oxidizing Archaea. Annual Review of Microbiology, 66, 83-101.https://doi.org/10.1146/annurev-micro-092611-150128
- 6. Lu, L. and Jia, Z. (2013) Urease Gene-Containing Archaea Dominate Autotrophic Ammonia Oxidation in Two Acid Soils. Environmental Microbiology, 15, 1795-1809.https://doi.org/10.1111/1462-2920.12071
- 7. Nicol, G.W., Leininger, S., Schleper, C. and Prosser, J.I. (2008) The Influence of Soil pH on the Diversity, Abundance and Transcriptional Activity of Ammonia Oxidizing Archaea and Bacteria. Environmental Microbiology, 10, 2966-2978.https://doi.org/10.1111/j.1462-2920.2008.01701.x
- 8. Zhang, L.-M., Hu, H.-W., Shen, J.-P. and He, J.-Z. (2012) Ammonia-Oxidizing Archaea Have More Important Role than Ammonia-Oxidizing Bacteria in Ammonia Oxidation of Strongly Acidic Soils. The ISME Journal, 6, 1032-1045. https://doi.org/10.1038/ismej.2011.168
- 9. Lu, L., Han, W., Zhang, J., Wu, Y., Wang, B., Lin, X., Zhu, J., Cai, Z. and Jia, Z. (2012) Nitrification of Archaeal Ammonia Oxidizers in Acid Soils Is Supported by Hydrolysis of Urea. ISME Journal, 6, 1978-1984.https://doi.org/10.1038/ismej.2012.45
- 10. Jia, Z.J. and Conrad, R. (2009) Bacteria rather than Archaea Dominate Microbial Ammonia Oxidation in an Agricultural Soil. Environmental Microbiology, 11, 1658-1671. https://doi.org/10.1111/j.1462-2920.2009.01891.x
- 11. Di, H.J., Cameron, K.C., Shen, J.P., Winefield, C.S., O’Callaghan, M., Bowatte, S. and He, J.Z. (2009) Nitrification Driven by Bacteria and Not Archaea in Nitrogen-Rich Grassland Soils. Nature Geoscience, 2, 621-624.https://doi.org/10.1038/ngeo613
- 12. Prosser, J.I. and Nicol, G.W. (2012) Archaeal and Bacterial Ammonia-Oxidisers in Soil: The Quest for Niche Specialisation and Differentiation. Trends in Microbiology, 20, 523-531. https://doi.org/10.1016/j.tim.2012.08.001
- 13. Pan, G., Li, L., Wu, L. and Zhang, X. (2004) Storage and Sequestration Potential of Topsoil Organic Carbon in China’s Paddy Soils. Global Change Biology, 10, 79-92.https://doi.org/10.1111/j.1365-2486.2003.00717.x
- 14. Park, B.J., Park, S.J., Yoon, D.N., Schouten, S., Damste, J.S.S. and Rhee, S.K. (2010) Cultivation of Autotrophic Ammonia-Oxidizing Archaea from Marine Sediments in Coculture with Sulfur-Oxidizing Bacteria. Applied and Environmental Microbiology, 76, 7575-7587. https://doi.org/10.1128/AEM.01478-10
- 15. Sauder, L.A., Albertsen, M., Engel, K., Schwarz, J., Nielsen, P.H., Wagner, M. and Neufeld, J.D. (2017) Cultivation and Characterization of Candidatus Nitrosocosmicus Exaquare, an Ammonia-Oxidizing Archaeon from a Municipal Wastewater Treatment System. The ISME Journal, 11, 1142-1157.https://doi.org/10.1038/ismej.2016.192
- 16. Chen, X., Zhang, L.M., Shen, J.P., Wei, W.X. and He, J.Z. (2011) Abundance and Community Structure of Ammonia-Oxidizing Archaea and Bacteria in an Acid Paddy Soil. Biology and Fertility of Soils, 47, 323-331.https://doi.org/10.1007/s00374-011-0542-8
- 17. Wei, X., Hu, Y., Peng, P., Zhu, Z., Atere, C.T., O’Donnell, A.G., Wu. J. and Ge, T. (2017) Effect of P Stoichiometry on the Abundance of Nitrogen-Cycle Genes in Phosphorus-Limited Paddy Soil. Biology and Fertility of Soils, 53, 767-776.https://doi.org/10.1007/s00374-017-1221-1
- 18. Ouyang, Y., Norton, J.M. and Stark, J.M. (2017) Ammonium Availability and Temperature Control Contributions of Ammonia Oxidizing Bacteria and Archaea to Nitrification in an Agricultural Soil. Soil Biology and Biochemistry, 113, 161-172.https://doi.org/10.1016/j.soilbio.2017.06.010
- 19. Stempfhuber, B., Richter-Heitmann, T., Bienek, L., Schöning, I., Schrumpf, M., Friedrich, M., Schulz, S. and Schloter, M. (2017) Soil pH and Plant Diversity Drive Co-Occurrence Patterns of Ammonia and Nitrite Oxidizer in Soils from Forest Ecosystems. Biology and Fertility of Soils, 53, 691-700.
- 20. Osborne, B.B., Baron, J.S. and Wallenstein, M.D. (2016) Moisture and Temperature Controls on Nitrification Differ among Ammonia Oxidizer Communities from Three Alpine Soil Habitats. Frontiers of Earth Science, 10, 1-12.https://doi.org/10.1007/s11707-015-0556-x
- 21. Delgado-Baquerizo, M., Maestre, F.T., Eldridge, D.J. and Singh, B.K. (2016) Microsite Differentiation Drives the Abundance of Soil Ammonia Oxidizing Bacteria along Aridity Gradients. Frontiers in Microbiology, 7, 505.
- 22. Muema, E.K., Cadisch, G. and Rasche, F. (2016) Soil Texture Modulates the Response of Ammonia-Oxidizing Prokaryotes to Biochemical Quality of Organic Inputs in Tropical Agricultural Soils. Soil Biology and Biochemistry, 100, 218-228.https://doi.org/10.1016/j.soilbio.2016.06.027
- 23. Lehtovirta-Morley, L.E., Stoecker, K., Vilcinskas, A., Prosser, J.I. and Nicol, G.W. (2011) Cultivation of an Obligate Acidophilic Ammonia Oxidizer from a Nitrifying Acid Soil. Proceedings of the National Academy of Sciences of the United States of America, 108, 15892-15897. https://doi.org/10.1073/pnas.1107196108
- 24. Koops, H.P. and Pommerening-Roser, A. (2001) Distribution and Ecophysiology of the Nitrifying Bacteria Emphasizing Cultured Species. FEMS Microbiology Ecology, 37, 1-9. https://doi.org/10.1111/j.1574-6941.2001.tb00847.x
- 25. Segal, L.M., Miller, D.N., McGhee, R.P., Loecke, T.D., Cook, K.L., Shapiro, C.A. and Drijber, R.A. (2017) Bacterial and Archaeal Ammonia Oxidizers Respond Differently to Long-Term Tillage and Fertilizer Management at a Continuous Maize Site. Soil and Tillage Research, 168, 110-117. https://doi.org/10.1016/j.still.2016.12.014
- 26. Hart, S.C., Stark. J.M., Davidson, E.A. and Firestone, M.K. (1994) Nitrogen Mineralization, Immobilization, and Nitrification. In: Methods of Soil Analysis: Part 2—Microbiological and Biochemical Properties, Soil Science Society of America, Madison, WI, 985-1018.
- 27. Rotthauwe, J.H., Witzel, K.P. and Liesack, W. (1997) The Ammonia Monooxygenase Structural Gene amoA as a Functional Marker: Molecular Fine-Scale Analysis of Natural Ammonia-Oxidizing Populations. Applied and Environmental Microbiology, 63, 4704-4712.
- 28. Tourna, M., Freitag, T.E., Nicol, G.W. and Prosser, J.I. (2008) Growth, Activity and Temperature Responses of Ammonia-Oxidizing Archaea and Bacteria in Soil Microcosms. Environmental Microbiology, 10, 1357-1364.https://doi.org/10.1111/j.1462-2920.2007.01563.x
- 29. Frijlink, M.J., Abee, T., Laanbroek, H.J., de Boer, W. and Konings, W.N. (1992) The Bioenergetics of Ammonia and Hydroxylamine Oxidation in Nitrosomonas europaea at Acid and Alkaline pH. Archives of Microbiology, 157, 194-199.https://doi.org/10.1007/BF00245290
- 30. Ju, X.-T., Xing, G.-X., Chen, X.-P., Zhang, S.-L., Zhang, L.-J., Liu, X.-J., Cui, Z.-L., Yin, B., Christie, P. and Zhu, Z.-L. (2009) Reducing Environmental Risk by Improving N Management in Intensive Chinese Agricultural Systems. Proceedings of the National Academy of Sciences of the United States of America, 106, 3041-3046.https://doi.org/10.1073/pnas.0813417106
- 31. Gao, S., Cao, W., Zou, C., Gao, J., Huang, J., Bai, J., Zeng, N., Shimizu, K.-Y., Wright, A. and Dou, F. (2017) Ammonia-Oxidizing Archaea Are More Sensitive than Ammonia-Oxidizing Bacteria to Long-Term Application of Green Manure in Red Paddy Soil. Applied Soil Ecology, in Press.https://doi.org/10.1016/j.apsoil.2017.09.041
- 32. Zhao, J., Wang, B. and Jia, Z. (2015) Phylogenetically Distinct Phylotypes Modulate Nitrification in a Paddy Soil. Applied and Environmental Microbiology, 81, 3218-3227. https://doi.org/10.1128/AEM.00426-15
- 33. Wang, J., Zhang, L., Lu, Q., Raza, W., Huang, Q. and Shen, Q. (2014) Ammonia Oxidizer Abundance in Paddy Soil Profile with Different Fertilizer Regimes. Applied Soil Ecology, 84, 38-44. https://doi.org/10.1016/j.apsoil.2014.06.009
- 34. Shen, J.P., Zhang, L.M, Zhu, Y.G., Zhang, J.B. and He, J.Z. (2008) Abundance and Composition of Ammoni-Oxidizing Bacteria and Ammonia-Oxidizing Archaea Communities of an Alkaline Sandy Loam. Environmental Microbiology, 10, 1601-1611.
- 35. Walker, C., De La Torre, J., Klotz, M., Urakawa, H., Pinel, N., Arp, D., Brochier-Armanet, C., Chain, P., Chan, P. and Gollabgir, A. (2010) Nitrosopumilus maritimus Genome Reveals Unique Mechanisms for Nitrification and Autotrophy in Globally Distributed Marine Crenarchaea. Proceedings of the National Academy of Sciences of the United States of America, 107, 8818-8823.https://doi.org/10.1073/pnas.0913533107
- 36. Chu, H., Fujii, T., Morimoto, S., Lin, X. and Yagi, K. (2008) Population Size and Specific Nitrification Potential of Soil Ammonia-Oxidizing Bacteria under Long-Term Fertilizer Management. Soil Biology and Biochemistry, 40, 1960-1963.https://doi.org/10.1016/j.soilbio.2008.01.006
- 37. Di, H., Cameron, K., Shen, J.P., Winefield, C., O’Callaghan, M., Bowatte, S. and He, J. (2009) Nitrification Driven by Bacteria and Not Archaea in Nitrogen-Rich Grassland Soils. Nature Geoscience, 2, 621-624. https://doi.org/10.1038/ngeo613
- 38. Li, H., Weng, B.S., Huang, F.Y., Su, J.Q. and Yang, X.R. (2015) pH Regulates Ammonia-Oxidizing Bacteria and Archaea in Paddy Soils in Southern China. Applied Microbiology and Biotechnology, 99, 6113-6123.https://doi.org/10.1007/s00253-015-6488-2
- 39. Pester, M., Rattei, T., Flechl, S., Gröngröft, A., Richter, A., Overmann, J., Reinhold-Hurek, B., Loy, A. and Wagner, M. (2012) amoA-Based Consensus Phylogeny of Ammonia-Oxidizing Archaea and Deep Sequencing of amoA Genes from Soils of Four Different Geographic Regions. Environmental Microbiology, 14, 525-539.https://doi.org/10.1111/j.1462-2920.2011.02666.x
- 40. Erguder, T.H., Boon, N., Wittebolle, L., Marzorati, M. and Verstraete, W. (2009) Environmental Factors Shaping the Ecological Niches of Ammonia-Oxidizing Archaea. FEMS Microbiology Reviews, 33, 855-869.https://doi.org/10.1111/j.1574-6976.2009.00179.x
- 41. Chen, X.P., Zhu, Y.G., Xia, Y., Shen, J.P. and He, J.Z. (2008) Ammonia-Oxidizing Archaea: Important Players in Paddy Rhizosphere Soil? Environmental Microbiology, 10, 1978-1987. https://doi.org/10.1111/j.1462-2920.2008.01613.x
- 42. Pett-Ridge, J., Petersen, D.G., Nuccio, E. and Firestone, M.K. (2013) Influence of Oxic/Anoxic Fluctuations on Ammonia Oxidizers and Nitrification Potential in a Wet Tropical Soil. FEMS Microbiology Ecology, 85, 179-194. https://doi.org/10.1111/1574-6941.12111
- 43. Konneke, M., Bernhard, A.E., de la Torre, J.R., Walker, C.B., Waterbury, J.B. and Stahl, D.A. (2005) Isolation of an Autotrophic Ammonia-Oxidizing Marine Archaeon. Nature, 437, 543-546. https://doi.org/10.1038/nature03911
- 44. Qin, W., Meinhardt, K.A., Moffett, J.W., Devol, A.H., Virginia Armbrust, E., Ingalls, A.E. and Stahl, D.A. (2017) Influence of Oxygen Availability on the Activities of Ammonia-Oxidizing Archaea. Environmental Microbiology Reports, 9, 250-256.https://doi.org/10.1111/1758-2229.12525
- 45. Wu, Y., Lu, L., Wang, B., Lin, X., Zhu, J., Cai, Z., Yan, X. and Jia, Z. (2010) Long-Term Field Fertilization Significantly Alters Community Structure of Ammonia-Oxidizing Bacteria rather than Archaea in a Paddy Soil. Soil Science Society of America Journal, 75, 1431-1439. https://doi.org/10.2136/sssaj2010.0434
- 46. Gubry-Rangin, C., Nicol, G.W. and Prosser, J.I. (2010) Archaea rather than Bacteria Control Nitrification in Two Agricultural Acidic Soils. FEMS Microbiology Ecology, 74, 566-574. https://doi.org/10.1111/j.1574-6941.2010.00971.x
- 47. Tourna, M., Stieglmeier, M., Spang, A., Könneke, M., Schintlmeister, A., Urich, T., Engel, M., Schloter, M., Wagner, M. and Richter, A. (2011) Nitrososphaera viennensis, an Ammonia Oxidizing Archaeon from Soil. Proceedings of the National Academy of Sciences of the United States of America, 108, 8420-8425.https://doi.org/10.1073/pnas.1013488108
- 48. Alam, M.S., Ren, G.D., Lu, L., Zheng, Y., Peng, X.H. and Jia, Z.J. (2013) Conversion of Upland to Paddy Field Specifically Alters the Community Structure of Archaeal Ammonia Oxidizers in an Acid Soil. Biogeosciences, 10, 5739-5753.https://doi.org/10.5194/bg-10-5739-2013
- 49. Wang, B., Zhao, J., Guo, Z., Ma, J., Xu, H. and Jia, Z. (2015) Differential Contributions of Ammonia Oxidizers and Nitrite Oxidizers to Nitrification in Four Paddy Soils. The ISME Journal, 9, 1062-1075. https://doi.org/10.1038/ismej.2014.194
- 50. Tourna, M., Freitag, T.E. and Prosser, J.I. (2010) Stable Isotope Probing Analysis of Interactions between Ammonia Oxidizers. Applied and Environmental Microbiology, 76, 2468-2477. https://doi.org/10.1128/AEM.01964-09
- 51. Bowatte, S., Jia, Z., Ishihara, R., Nakajima, Y., Asakawa, S. and Kimura, M. (2006) Molecular Analysis of the Ammonia Oxidizing Bacterial Community in the Surface Soil Layer of a Japanese Paddy Field. Soil Science & Plant Nutrition, 52, 427-431. https://doi.org/10.1111/j.1747-0765.2006.00058.x
- 52. Avrahami, S. and Conrad, R. (2003) Patterns of Community Change among Ammonia Oxidizers in Meadow Soils upon Long-Term Incubation at Different Temperatures. Applied and Environmental Microbiology, 69, 6152-6164.https://doi.org/10.1128/AEM.69.10.6152-6164.2003
- 53. Wang, Y., Ke, X., Wu, L. and Lu, Y. (2009) Community Composition of Ammonia-Oxidizing Bacteria and Archaea in Rice Field Soil as Affected by Nitrogen Fertilization. Systematic and Applied Microbiology, 32, 27-36.https://doi.org/10.1016/j.syapm.2008.09.007
- 54. Burton, S.A. and Prosser, J.I. (2001) Autotrophic Ammonia Oxidation at Low pH through Urea Hydrolysis. Applied and Environmental Microbiology, 67, 2952-2957.https://doi.org/10.1128/AEM.67.7.2952-2957.2001
- 55. Zhao, W., Cai, Z.-C. and Xu, Z.-H. (2007) Does Ammonium-Based N Addition Influence Nitrification and Acidification in Humid Subtropical Soils of China? Plant and Soil, 297, 213-221. https://doi.org/10.1007/s11104-007-9334-1
- 56. Zhao, W., Zhang, J., Müller, C. and Cai, Z. (2017) Mechanisms behind the Stimulation of Nitrification by N Input in Subtropical Acid Forest Soil. Journal of Soils and Sediments, 17, 2338-2345. https://doi.org/10.1007/s11368-016-1461-y
Supplementary
Figure 1S. The copy number of bacterial 16S rRNA genes in the eight different paddy soils. The error bars represent the standard errors of the means of the triplicates. All the designations are the same as those in Table 1.
Table S1. Sampling site characteristics.
Table S2. Primers and conditions used in this study.
The forward primer of bacterial amoA amoA-1F was attached with a 27 bp GC clamp (Nicolaisen and Ramsing 2002 [4] ).
Supplementary Reference
[1] Stubner, S. (2002) Enumeration of 16S rDNA of Desulfotomaculum Lineage 1 in Rice Field Soil by Real-Time PCR with SybrGreen™ Detection. Journal of Microbiological Methods, 50, 155-164. https://doi.org/10.1016/S0167-7012(02)00024-6
[2] Tourna, M., Freitag, T.E., Nicol, G.W. and Prosser, J.I. (2008) Growth, Activity and Temperature Responses of Ammonia-Oxidizing Archaea and Bacteria in Soil Microcosms. Environmental Microbiology, 10, 1357-1364. https://doi.org/10.1111/j.1462-2920.2007.01563.x
[3] Avrahami, S., Liesack, W. and Conrad, R. (2003) Effects of Temperature and Fertilizer on Activity and Community Structure of Soil Ammonia Oxidizers. Environmental Microbiology, 5, 691-705. https://doi.org/10.1046/j.1462-2920.2003.00457.x
[4] Nicolaisen, M.H. and Ramsing, N.B. (2002) Denaturing Gradient Gel Electrophoresis (DGGE) Approaches to Study the Diversity of Ammonia-Oxidizing Bacteria. Journal of Microbiological Methods, 50, 189-203. https://doi.org/10.1016/S0167-7012(02)00026-X
NOTES
#These authors contributed equally to this work.