Functional Data Analysis of Spectroscopic Data with Application to Classification of Colon Polyps ()
1. Introduction
Near-infrared (NIR) spectra of biomedical objects consist of many overlapping absorption bands representing the different modes of vibration of a large number of molecular constituents in the compounds, which are sensitive to the physical and chemical states of the compounds [1] [2] Spectroscopic data show highly correlated structure due to the complex nature of its spectral absorption bands. The underlying information thus varies smoothly with spectral wavelength. This feature makes spectral data different from the typical statistical high-dimensional data.
Traditional spectral data analysis involves multivariate statistical techniques such as multiple linear regression, principal components regression (PCR) [3] [4] and partial least squares (PLS) regression [5] [6] . However, these methods consider the spectrum as a set of discrete variable points rather than as a conti- nuous function. From a physical point of view it would be more informative to describe the spectrum as a smooth function, being a sum of absorption peaks caused by the different chemical constituents present in the sample under study, where the absorbance at two wavelengths close to each other is shown highly correlated.
Functional data analysis (FDA) proposed by Ramsay and Silverman [7] is used to deal with data of a functional nature, where each spectrum is replaced by its approximation by a linear combination of smooth basis functions. In recent years, there have been an increasing number of publications on FDA, including functional linear regression with functional predictors and scalar responses, and its applications in NIR spectroscopy, particularly in the area of biomedical applications [8] [9] [10] . In classification problems, generalized linear model was adapted to the presence of functional predictor variables [11] [12] . To deal with the problem of multicollinearity in the multiple linear regression of functional data, functional principal component analysis and functional partial least squares were introduced to the classification problem of functional data [13] .
In this study of functional logistic regression (FLR) [14] models with different functional basis were explored to distinguish the differences between the two polyps in the colon for detection of precancerous adenomatous polyps. PCA basis and PLS basis [7] were investigated and insights were gained on the effects of different functional representations of predictors. The multivariate methods using principal component discriminant analysis (PCDA) and partial least squares discriminant analysis (PLSDA) were also explored for classification. The performances of the FLR models were compared with that of the PCDA and PLSDA models in terms of both classification accuracy and understanding of spectral features.
2. Materials and Methods
2.1. Spectroscopic Data Description
A colon dataset is presented here to illustrate the application of functional data analysis in a classification setting with a problem arising in colonoscopy. The evolution of colon cancer starts with colon polyps. There are two different types of colon polyps, hyperplastic and adenomatous polyps. Hyperplasias are benign polyps which are known not to evolve into cancer. Adenomas have a strong tendency to develop cancer and therefore they have to be excised immediately. Polyps are often found during endoscopy of the colon (colonoscopy). A method to differentiate reliably adenomas from hyperplasias during a preventive medical colonoscopy is highly desirable.
Visible-NIR spectra covering the range from 320 to 920 nm were collected from 64 human colonic biopsy sites using an optical probe in vivo during colonoscopy which was undertaken to assess patients for precancerous adenomatous polyps. Repeated measurements, on average 6 spectra per site, were taken from each biopsy site. In total 363 spectra were analyzed, including 63 spectra from 11 hyperplastic polyps and 300 spectra from 53 adenomatous polyps.
2.2. Spectral Pre-Treatment
Before constructing a classification model, standard data pre-processing was carried out on the spectra to improve signal quality. This involved spectral smoothing using Savitzky-Golay method (Savitzky and Golay 1964) [15] , cropping the noisy ends of the spectra, and normalizing using the standard normal variate (SNV) method [16] . After pretreatment, only the spectra in the range of 370 - 800 nm were used in our analysis. Spectra from the two types of polyps are of very similar overall shape, with the mean spectra showing small between-class differences as shown in Figure 1.
2.3. Statistical Analysis
2.3.1. Principal Component Discriminant Analysis (PCDA) and Partial Least Squares Discriminant Analysis (PLSDA)
Multivariate statistical methods including principal component analysis (PCA), partial least squares (PLS) and linear discriminant analysis (LDA) [17] [18] [19] were employed to investigate the differences of spectra from hyperplastic and adenomatous polyps. Direct implementation of LDA in high-dimensional spectroscopic data setting provides poor classification results and the interpretation of the results is challenging due to singularity problem and highly-correlated spectral features. To solve this problem, PCA or PLS was used to construct a reduced number of new uncorrelated variable with maximal variance or maximal covariance with response. Then LDA focuses on finding a linear combination of the new variables to construct canonical variate which gives the maximum separation of the two groups. Using pretreated spectral data described in Section 2.2, classification rules were derived using principal component discriminant analysis (PCDA) [20] [21] [22] and partial least squares discriminant analysis (PLSDA) [20] [21] [22] [23] . The PCDA involves an initial PCA on the pre-treated spectra followed by a LDA performed on the first k PCs’ scores. The PLSDA involves a PLS regression on the pre-treated spectral data using as response variable y a dummy variable, coded as 0 or 1 for spectrum from hyperplastic and adenomatous polyps respectively. This data reduction step is followed by a LDA on the first k PLS components’ scores. Both PCDA and PLSDA were carried out with k ranging from 2 to 30 and the number of components k was optimized by using cross-validation. The leave-out-one-site cross-validation was used in this work to train the algorithm by carrying out the classification rules on all the data except one biopsy site which was then tested. This was repeated until all sites have been tested and an overall model accuracy was determined. This was used to assess the performance of the classification as measured by sensitivity and specificity.
2.3.2. Functional PCA and Functional PLS Regression
Though multivariate PCA and PLS are powerful exploratory statistical methods to reduce the dimension of the original spectral variable, in spectroscopic data setting it would be more informative to describe the spectrum as a smooth function rather than as a set of points when taking into account the underlying spectral information.
To solve the problems of high dimensionality and multicollinearity encountered in functional regression models, the PCA and PLS have been generalized to the infinite-dimensional case where observations of predictor variables are cur- ves or functions (functional data) instead of vectors as in multivariate case. Functional PCA [7] [13] and Functional PLS [7] [8] [13] [14] are useful tools for studying functional data by providing common functional components so as to analyze the variability of functional data in an understandable manner. By approximating infinite-dimensional random functions by a finite number of random score vectors, functional PCA appears as a dimension reduction technique just as in the multivariate case and thus reduces the complexity of the spectroscopic data.
Let  be a random sample of observations (sample paths) of a functional predictor variable
be a random sample of observations (sample paths) of a functional predictor variable 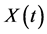 where
where 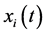 is the functional predictor of i th spectrum with t the function argument. In this study, T is the spectral wavelength interval extending from 370 nm to 800 nm. Let
is the functional predictor of i th spectrum with t the function argument. In this study, T is the spectral wavelength interval extending from 370 nm to 800 nm. Let  be a set of observations of a binary response variable Y where each
be a set of observations of a binary response variable Y where each 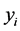 is a dummy variable, coded as either 0 or 1 for the i th spectrum from either hyperplastic or adenomatous polyps respectively. We assume that the random variables X and Y are defined on the same probability space with X valued in the space
is a dummy variable, coded as either 0 or 1 for the i th spectrum from either hyperplastic or adenomatous polyps respectively. We assume that the random variables X and Y are defined on the same probability space with X valued in the space 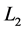 of square integrable functions on T.
of square integrable functions on T.
Functional PC and functional PLS components are uncorrelated that are obtained as generalized linear combinations of the functional predictor variables. The j th component is defined by
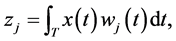 (1)
(1)
where 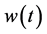 are weight functions that are the solutions to the following optimization problems.
are weight functions that are the solutions to the following optimization problems.
For functional PCA, we seek weight functions 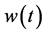 such that the variance of components defined in Equation (1) is maximal.
such that the variance of components defined in Equation (1) is maximal.
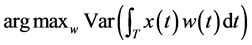 (2)
(2)
subject to 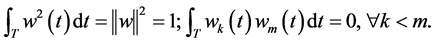
For functional PLS, we seek weight functions  such that the covariance between the response and latent components constructed in Equation (1) is ma- ximal.
such that the covariance between the response and latent components constructed in Equation (1) is ma- ximal.
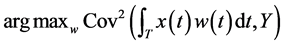 subject to
subject to 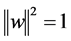 (3)
(3)
2.3.3. Functional Logistic Regression (FLR) with Functional PC Basis and Functional PLS Basis
In binary classification problems, a functional logistic regression (FLR) model is used to model the relationship between the binary response and the functional predictor whose observations are functions instead of vectors as in multivariate case. The FLR model is given by
 (4)
(4)
where , the probability that the binary response variable takes value one given a functional observation, is modeled as
, the probability that the binary response variable takes value one given a functional observation, is modeled as
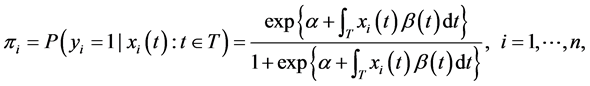 (5)
(5)
with  being a real parameter,
being a real parameter, ![]() the functional parameter that belongs to the space
the functional parameter that belongs to the space![]() , and
, and ![]() are independent random errors.
are independent random errors.
Taking the logit transformation gives the following equivalent expression for FLR model:
![]() (6)
(6)
The use of least squares criteria to estimate the parameters of the functional linear regression model yields an ill-posed problem due to the infinite dimension of the predictor space. In addition, sample path ![]() are measured discretely. The most common solution to this problem is to assume that both the sample paths
are measured discretely. The most common solution to this problem is to assume that both the sample paths ![]() and the parameter function
and the parameter function ![]() belong to a finite dimension space spanned by a basis of functions. We then have following representations of
belong to a finite dimension space spanned by a basis of functions. We then have following representations of ![]() and
and ![]() as a linear combination of the basis functions
as a linear combination of the basis functions![]() :
:
![]() (7)
(7)
where ![]() is a vector of basis functions,
is a vector of basis functions, ![]() is the vector of sample path basis coefficients and
is the vector of sample path basis coefficients and ![]() is the parameter function basis coefficients. Therefore, the functional linear regression model is equivalent to the multiple linear model
is the parameter function basis coefficients. Therefore, the functional linear regression model is equivalent to the multiple linear model
![]() (8)
(8)
where ![]() and
and
![]() ,
, ![]()
The main problem is to approximate the basis coefficients of sample curves from their discrete observations and to select an appropriate basis by taking into account the characteristics of the observed sample curves. Due to the high dependence structure of the design matrix ![]() in the model, the multiple linear regression model shows high multicollinearity. Functional PCA and functional PLS approaches as described in Section 2.3.2 were used here so as to construct a reduced set of uncorrelated components of the design matrix. The problem is then reduced to regressing a logit transformation of response variable on an optimum set of orthogonal (principal or PLS) components obtained as generalized linear combinations of the functional predictor that solve optimization criteria in Equations (2) and (3). In the rest part of this paper, FPCA and FPLS are used to refer to functional PCA logistic regression model and functional PLS logistic regression model, respectively.
in the model, the multiple linear regression model shows high multicollinearity. Functional PCA and functional PLS approaches as described in Section 2.3.2 were used here so as to construct a reduced set of uncorrelated components of the design matrix. The problem is then reduced to regressing a logit transformation of response variable on an optimum set of orthogonal (principal or PLS) components obtained as generalized linear combinations of the functional predictor that solve optimization criteria in Equations (2) and (3). In the rest part of this paper, FPCA and FPLS are used to refer to functional PCA logistic regression model and functional PLS logistic regression model, respectively.
The FPCA and FPLS were carried out with k, the number of component basis functions, ranging from 2 to 30 and k was optimized by using the leave-out-one- site cross-validation as mentioned in Section 2.3.1.
All the algorithms for computations and analyses were implemented in R statistical programming language [25] using the R packages MASS and fda.usc [26] .
3. Results and Discussion
The mean spectral patterns from hyperplastic and adenomatous polyps after standard pre-treatment are shown in Figure 1. It illustrates that the spectra of the two types of polyps are of very similar overall shape, with the mean spectra showing small between-class differences.
3.1. Discrimination by PCDA and PLSDA Models
For PCDA and PLSDA models, the discrimination results of cross-validation were used to optimize the number of PCs or PLS components. In Table 1, the PCDA model constructed on the first twenty PCs gave the best discrimination accuracy with sensitivity and specificity of 81% and 76%, respectively. When
![]()
Figure 1. Mean spectra (± one standard deviation) from hyperplastic (solid blue line for mean and dotted blue line for standard deviation) and adenomatous (dashed red line and dotted red lines for standard deviation) polyps of colon data after standard pre-treatment.
taking into account the relationship between the spectral variables and the response variable for latent variable design, the optimal PLSDA model achieved a leave-one-out cross-validation discrimination accuracy with the same sensitivity and specificity of 81% and 76% respectively, but using only seven PLS components in constructing the canonical variable.
By combining the loadings from the PCA or PLS and LDA, the PCDA loading and PLSDA loading can be found to show the contribution at each wavelength to the linear discrimination rule and thus can be related easily to the spectral features for interpretation purpose.
However, in Figure 2 the loadings from these two models look complex, especially for the PCDA model with more number of components used for the discrimination, thus the contribution of each wavelength to the classification becomes less interpretable when it is related to the spectral features.
3.2. Discrimination by FPCA and FPLS Logistic Regression Models
Though the PLSDA model used a smaller number of constructed variables for classification when compared with the PCDA model, the classification perfor- mance and interpretation of both models are not satisfactory. FPCA and FPLS logistic regression models showed improvements over the multivariate methods.
![]()
Table 1. Classification results of different models using leave-one-out cross-validation for differentiating between hyperplastic and adenomatous polyps of colon data.
![]()
Figure 2. PCDA loadings (left panel) and PLSDA loadings (right panel) for discrimination between hyperplastic and adeno- matous polyps of colon data. The discrimination loading is shown in green, with the mean spectra for the two types superimposed (hyperplastic solid blue line, adenomatous dashed red line).
The discrimination results of cross-validation were used to optimize the num- ber of PCs or PLS components basis functions. In Table 1, the FPCA models with twelve functional PCs gave the optimal results, with sensitivity and specificity of 88% and 76% respectively. The FPLS models with seven functional PLS components gave the optimal results, with both sensitivity and specificity of 85% and 76% respectively. Here a specificity of 76% was chosen considering the small size of colon data so as to make sensitivity comparable for different models. As shown in Figure 3, when we compare the FPCA models with the PCDA models having the same number of components, for the models using more than ten PCs, the FPCA model can often achieve higher classification accuracy than PCDA model. When we compare the FPLS models with the PLSDA models having the same number of components, for the models using at least seven PLS components, the FPLS model can often achieve higher classification accuracy than PLSDA model.
Using a small number of PC or PLS functional basis components, the coefficients of FPCA and FPLS models show simpler structure features. This allows the visualization of discrimination coefficient vectors and showed how they contribute to the correct classification clearly. In Figure 4, the obvious features with large positive loadings from both FPCA and FPLS models concentrated at around 410 nm, 540 nm and 580 nm, corresponding to some small differences between the mean spectra of hyperplastic and adenomatous polyps. The effect of absorption seems to play a key role in the spectral differences and the classification between two types of tissue in some specific spectral region. These informative regions might be due to absorption dips of HbO2 at 415 nm, 542 nm and 577 nm in the spectra of high risk cancer due to increased Hb concentration [27] . It is known that cancers and pre-cancerous tissues are characterized by increased microvascular volume, hence increased blood content [28] .
![]()
Figure 3. Left panel: FPCA and PCDA classification accuracy of colon data as measured by sensitivity; right panel: FPLS and PLSDA classification accuracy of colon data as measured by sensitivity.
We further explored the structures of the FPCA and FPLS components. The first four functional PCs were considered since they account for more than 95% of the total variation in the colon spectral data. As shown in Figure 5, the first and dominant component is simple in structure and resembles a single cycle of a sine wave. The first two functional PCs with large positive or negative loadings around 410 nm, 540 nm and 580 nm explain the informative spectral features in these regions. The subdominant components are roughly sinusoidal, but show
![]()
Figure 4. FPCA loadings (left panel) and FPLS loadings (right panel) for discrimination between hyperplastic and adenomatous polyps of colon data. The functional regression loading is shown in green, with the mean spectra for the two types superimposed (hyperplastic solid blue line, adenomatous dashed red line).
![]()
Figure 5. The first four functional PC loadings (top panel) and the first four functional PLS loadings (bottom panel) from colon data. Functional PC loading and functional PLS loading are shown in green line, with the mean spectra for the two types of polyps superimposed (hyperplastic in solid blue line, adenomatous in dashed red line).
more and more cycles. The functional PLS loadings seem to capture more informative spectral features even for dominant component.
4. Conclusions
Functional data analysis has been widely used in spectroscopic data where the absorbance spectrum is a functional variable whose observations are functions of wavelength. In this study, two functional logistic regression models with functional PC basis and functional PLS basis have been developed to distinguish adenomatic polyps from hyperplastic polyps during endoscopy of the colon. The results of this study showed that the functional logistic regression models outperformed the PCDA and PLSDA models by using a small number of components.
The commonly used multivariate models PCDA and PLSDA with more discriminant components used in the models may include some noise to hinder the classification performance and the interpretation of the spectral feature may be challenging. Taking into account the functional form of spectroscopic data, the FPCA and FPLS logistic regression models improved classification accuracy. The functional representation of the spectra combines dimension reduction and smoothing in one step. Both the FPCA and FPLS models gave better classification performance than the PCDA and PLSDA models and used a reduced number of functional basis components. In particular, FPLS used fewer latent variables than FPCA.
The most important contribution of the FPCA and FPLS models is that the discrimination coefficients that contributed to the correct classification of hyper- plastic and adenomatic polyps provided us insights and good understanding of the complex spectral features related to different types of colon polyps. With substantial reduction of spectral components, this functional logistic regression model is also a potentially accurate, fast and robust tool to distinguish adenomatic polyps from hyperplastic polyps during endoscopy of colon for the detection and removal of precancerous polyps before they turn into cancer. This is crucial for real-time clinical diagnostic application. In the future it is of interest to test the models on a larger number of colonic polys samples to give a more reliable diagnostic result. The spectral feature selection in functional data classification and interpretation of the classification loadings will also be further developed in future work.
Acknowledgements
This research is funded by Academic Research Funds (AcRF: RI 6/14 ZY) of National Institute of Education, Nanyang Technological University, Singapore. The author thanks the National Medical Laser Centre of University College London for providing the vis-NIR spectroscopic data.