Non-Stem Amino Acids Are Involved in the Phage P22 TSP NTD Stability ()
1. Introduction
The bacteriophage (phage) P22 is a well-studied phage system as well as being an intensely used model for a number of life processes and proteins. P22 infects a very small number of Salmonella species, including Salmonella typhimurium. Its assembly pathway has been studied in great detail [1] -[3] . Cryoelectron microscopy has been used in recent years to describe the protein structure of phage particles in near atomic detail [4] [5] . The P22 phage is a member of the Podoviridae family of bacteriophages, having a short non-contractile phage tail structure [6] .
The P22 tailspike protein (TSP) is one of the most well-studied phage proteins. It is the last protein to be added in the phage assembly pathway to yield a completed infectious particle [7] [8] . This last assembly step, when done in vitro, is called tailing. The P22 TSP, the subject of this study, is a trimeric protein consisting of 666aa in which the first stable form of the tail protein is its trimeric form [9] [10] . In addition to its pivotal assembly role, the P22 TSP is responsible for selection and attachment of the phage to the bacterial host cell lipopolysaccharide (LPS) receptor [11] .
The P22 TSP has several novel characteristics: resistance to sodium dodecyl sulfate (SDS) denaturation, resistant to heat (Tm of 88.3˚C) and resistance to proteolysis [12] [13] . Each chain consists of three domains: the N-terminal domain (NTD, aa1-aa108), the β-helix (aa143-aa540) and the trimerization domain (aa541-aa666; see Figure 1). The P22 TSP NTD binds to the crevice formed between the GP4 and GP10 viral structural proteins while a crevice formed by two sets of peptide ridges, located in the β-helix domain is responsible for host cell LPS binding and hydrolysis [14] [15] . Recent studies on the structure and function of the P22 TSP NTD have found amino acids, located within the interior of the NTD dome, are involved in the stability of the domain structure [16] . This Short Communication presents preliminary studies strongly suggesting two amino acids, which are not localized in the stem region, and may be important determinants in the overall stability of the P22 TSP NTD.
2. Materials and Methods
2.1. Primers and Mutagenesis
The two tail mutations, studied in this Short Communication, were isolated by the Berget Laboratory [17] . They were studied here because they were the only two mutations found to be located in the P22 TSP NTD [17] . The P22 TSP NTD domain is an important research focus of this laboratory. They were: hmH4-1 (G29R; GGG < codon 29 > AGG) and hmH3 (G71D; GGT < codon 71 > GAT). The following primers were used to transfer the Berget NTD mutations onto pET11a-TSP. The primers for the mutation hmH4-1 were: 5-G28R (or: aaagctgttgctaataggaaaatttacattggt) and 3-G28R (or: ac caatgtaaattttcctattagcaacagctt). The primers for the muta-
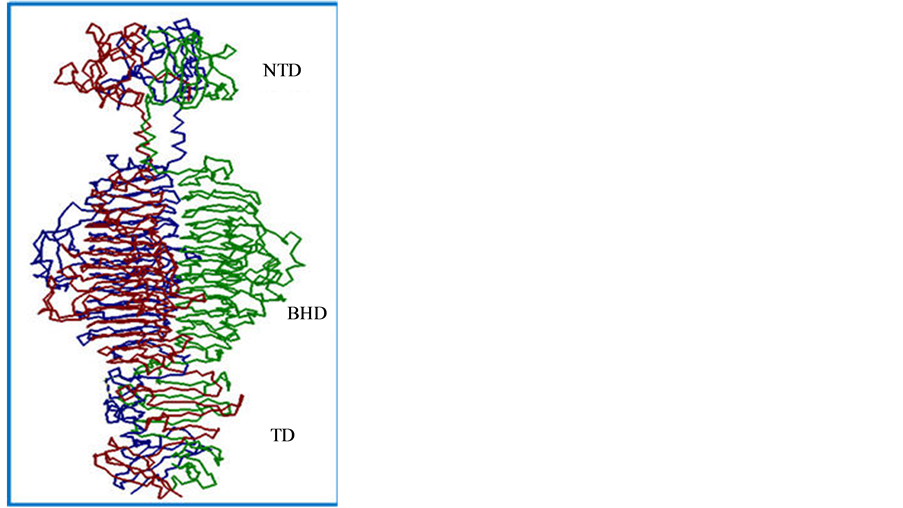
Figure 1. Trimeric structure of the P22 tailspike protein. The three domains of the P22 TSP are: the N-terminal domain (NTD), the beta-helix domain (BHD) and the trimerization domain (TD).
tion hmH3 were: 5G70D (or: attatcaacgcagccgataaaatcgtatacaac) and 3G70D (or: gttgtatacgattttatcggctgcgttgataat). The tail mutations were transferred from primers to the pET11a-TSP DNA by the StratagenQuikChange Mutagenesis protocol. The E. coli strain BL21/DE3, used for gene expression, was from Novagen.
2.2. Biological and Recombinant Protocols
The biological, recombinant and expression protocols have been described [16] [18] . The expression plasmid, pET11a-TSP containing a wild type copy of the P22 tailspike gene, was from Dr Peter Prevelige (University of Alabama at Birmingham, UAB). The protein function of the expressed cloned tail gene or gene variant was determined by the Berget Assay [16] . The Berget Assay is a high-expression variant of the functional petri plate assay devised by Schwarz and Berget [17] . It has been described in detail recently [16] .
3. Results and Discussion
3.1. Mutations in the DNA Sequence Corresponding to the N-Terminal Domain
Two previously identified mutations were localized to the gene region corresponding to the NTD. These mutations hmH4-1 and hmH3 were transferred to pET11a-TSP by the QuikChange Mutagenesis protocol and plasmid mutational sequences were confirmed by DNA sequencing using the sequencing facilities of the University of Alabama at Birmingham (UAB Heflin Genome Sequencing Facility).
3.2. Assay of Cloned Tail Protein Function
The functional nature of hmH4-1 (pET11a-TSP-G29R), hmH3 (pET11a-TSP-G71D) and the wild type tail gene (pET11a-TSP) was determined by the Berget Assay [16] . This test measures but does not distinguish between all three tail activities: binding to the viral particle (NTD function), binding LPS and LPS hydrolysis by tail endorhamnosidase activity (both functions of the β-helix domain). By this simple yet extremely sensitive petri plate assay these tail variants had no detectable tail function while the wild type tail gene, carried on plasmid, pET11a-TSP, had tail activity. These data confirmed the previous determination of the tail function of these two tail mutations [17] . The amino acid substitutions at amino acid positions aa29 and aa71 (out of 666aa) were both localized to the surface of the NTD [15] . To determine if the mutational inactivation of tail function could be due to structural alteration, a SDS-PAGE analysis was carried out as described in the following section.
3.3. SDS PAGE Analysis
Protein separation by SDS-PAGE normally results in denaturation of the applied proteins and their separation according to size. If the protein sample is heated prior to application to SDS PAGE, all protein will migrate according to their molecular size with the fastest migration occurring with the smaller proteins. However, there are a very small number of proteins that are refractory to SDS denaturation (without prior heating), including the P22 TSP [19] . The P22 TSP migrates as a trimeric protein with no prior heating [13] . However, there is another example of altered tailspike electrophoretic migration. Tailspike mutants have been found that destabilize the NTD and a thermal intermediate has been identified both of which destabilize the NTD resulting in the same migration which is slightly faster than the unheated trimeric TSP [16] [20] [21] .
Thus the fully denatured, the NTD-denatured and wild type TSP conformations can be distinguished via electrophoretic migration study in SDS-PAGE with unheated samples. Such a study has been carried out and is depicted in Figure 2 and Figure 3. Figure 2 displays the partially purified extracts of WT TSP and TSP-G29K mutant protein while the gel figure (Figure 3) displays the partially purified extracts of WT TSP and TSP-G71D mutant protein. In both gels the WT TSP migrates above the 180 kDa marker (TSP is 210 kDa). In both cases, the fully denatured TSP migrates below the 90 kDa marker (TSP is 72 kDa monomer). The unheated forms of the TSP-G29K and TSP-G71D (Figure 2, Lane 5 and Figure 3, Lane 4 respectively) migrate below the WT TSP but well above their denatured forms (Figure 2, Lane 3 and Figure 3, Lane 2 respectively). This is the location of the previously identified TSP forms whose NTD was denatured but not the other domains.
4. Discussion
The two sites G29K and G71D caused the N-terminal domain of the P22 TSP to collapse. Future studies will in-
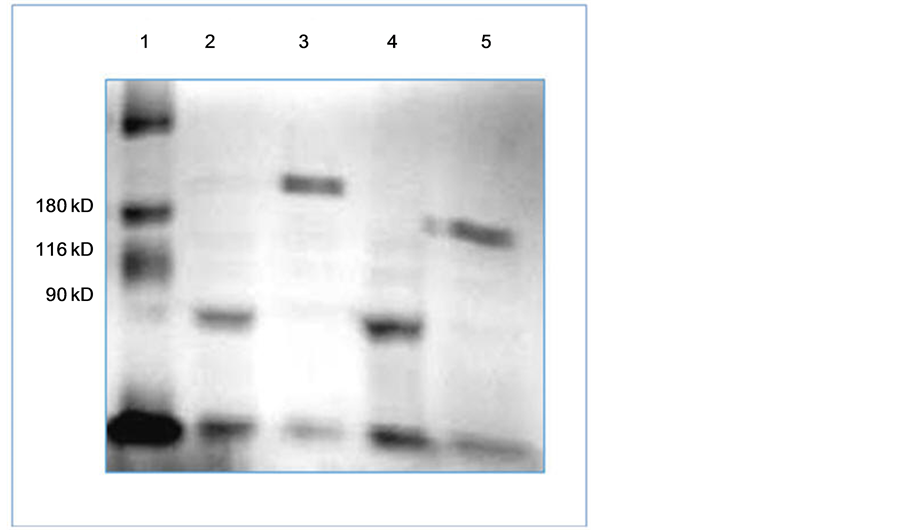
Figure 2. SDS-PAGE analysis of TSP-G29R. The gel contains Broad Range MW markers (Lane 1, L1), WT TSP (L2, L3), and TSP-G29R, (L4, L5) both heated and unheated.

Figure 3. SDS-PAGE analysis of TSP-G71D. The gel contains Broad Range MW markers (Lane 1, L1), WT TSP (L2, L3), and TSP-G71D, (L4, L5) both unheated and heated.
clude the determination of the nature of the two sites. It will be determined if these sites are tolerant to amino acid substitutions and the nature of the tolerated substitutions. Suppressor analyses will be carried out to determine if these two sites interact with other sites (intragenic suppressor analyses) in order to stabilize the P22 NTD.
5. Conclusion
This study strongly suggests that the amino acids at positions aa29 and aa71 are critical for the stability of the trimeric dome-like structure of the N-terminal domain. Further studies are in progress.
Acknowledgements
The authors thank Drs Doba Jackson (Huntingdon College), B. K. Robertson (Alabama State University) and H. Wu (Alabama State University) for discussion. This study is partially supported by funds from Department of Biological Sciences of the College of Science, Mathematics and Technology and the Program in Microbiology of the Alabama State University.
NOTES
*Corresponding author.