1. Introduction
Cancer grows if
and
(1)
and is eliminated if the reverse inequality holds. Here
and
are initial conditions in
, see section three for definitions and also [1] . So if you have many (few) growth inhibitors compared to growth factors, cancer is eliminated (cancer grows).
In [1] we proved, that Formula (1) when
implied that cancer grows and is eliminated if the reverse inequality holds. In the present paper we prove, that cancer grows if
and
(2)
and is eliminated if the reverse inequality holds. In [1] we also considered a mass action kinetic system with vector field f like the one in Section 4 with
and proved, that there is a relationship between such a model and the model T of Section 3. Namely if you linearize f at a singular point and then discretize the flow then you get a mapping T of Section 3. See section 4 for details.
Consider now the cancer model from [1]
(3)
Here
, where T denotes a transpose. If you fit my model to measurements, you will get some information about the particular cancer.
is the cancer agressiveness parameter. If this parameter is high cancer initially proliferates rapidly.
is the carcinogen severity.
is the fitness of the immune system, its response to cancer.
are decay rates. g is a vector of birth rates.
gives the growth factor response to cancer and
gives the growth inhibitor response to cancer. So fitting my model may have prognostic and diagnostic value. If we have a toxicology constraint for chemo therapy or immune therapy with a suitable safety margin
(4)
then we can keep the system at the toxicology limit by requiring
(5)
which is equivalent to
(6)
If
, then we can give chemo therapy at this rate. Then we get the induced system
(7)
(8)
We shall prove that this treatment benefits the patient in section 2. To get the system to the toxicology limit P assume that we have
(9)
Then looking at the third coordinate of T we see that we shall require
(10)
which implies that
(11)
We can also fit the ODE model of section 4 with
, by defining the Euler map
(12)
. Iterating this map will give an approximation to the flow. Then
is the rate at which you give chemo therapy. If we have the constraint
(13)
then looking at the third coordinate of H we see that to keep the system at the toxicology limit with a suitable safety margin, we must have
(14)
Solving for
we get
(15)
Since the
are positive we can give the chemo therapy at this rate. To get this system to the toxicology limit we shall require
(16)
which means, that
(17)
I felt I had to suggest this. If you want to try this you may want to do it stepwise.
In Figure 1, I have plotted a fit of T to three Gompertz functions
(18)
![]()
Figure 1. A fit to Gompertz functions. The upper curve is
, the middle
and the lower curve is
. The solid curves are the Gompertz functions and the dots the model T.
(19)
(20)
From a paper from 1964 [2] we know that solid tumors grow like Gompertz functions. That is the cancer burden is approximately a Gompertz function. Define the error functions
(21)
(22)
(23)
where
are measurements of
at equidistant time points
. We set
,
,
Then solve the equations
(24)
(25)
(26)
(27)
in unknowns
and
(28)
(29)
(30)
in unknowns
and
(31)
(32)
(33)
in unknowns
. For instance
(34)
gives
(35)
The result is
(36)
(37)
(38)
(39)
(40)
(41)
(42)
(43)
(44)
(45)
I have also fitted S to two Gompertz functions
(46)
(47)
See Figure 2 and Figure 3. Define error functions
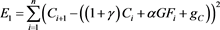 (48)
(48)
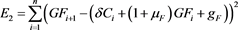 (49)
(49)
![]()
Figure 2. A fit of S to a Gompertz functions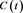 . The solid curve is the Gompertz function and the dots are the model S.
. The solid curve is the Gompertz function and the dots are the model S.
![]()
Figure 3. A fit of S to a Gompertz functions . The solid curve is the Gompertz function and the dots are the model S.
. The solid curve is the Gompertz function and the dots are the model S.
and measurements . Solve
. Solve
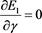 (50)
(50)
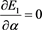 (51)
(51)
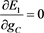 (52)
(52)
in unknowns 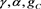 and
and
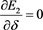 (53)
(53)
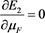 (54)
(54)
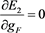 (55)
(55)
in unknowns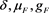 . The result is
. The result is
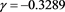 (56)
(56)
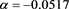 (57)
(57)
![]() (58)
(58)
![]() (59)
(59)
![]() (60)
(60)
![]() (61)
(61)
In Maple, there is a command QPSolve that minimizes a quadratic error function with constraints on the signs of the parameters estimated. There are several important monographs relevant to the present paper, see [3] - [8] . There are several publications by the author impacting on the present paper, see [9] - [15] .
2. The Routh Hurwitz Criterion for Maps
We shall derive a well known criterion for stability of a fixed point of a map. To this end define the Möbius transformation
![]() (62)
(62)
which maps the left hand plane ![]() to the interior
to the interior ![]() of the unit disc. This is because
of the unit disc. This is because
![]() (63)
(63)
implies
![]() (64)
(64)
![]() , and
, and
![]() (65)
(65)
implies
![]() (66)
(66)
Define
![]() (67)
(67)
Then
![]() (68)
(68)
when ![]() lies in the interior of
lies in the interior of![]() . Also
. Also
![]() (69)
(69)
![]() (70)
(70)
This shows, that g is a bijective map with inverse![]() . Let
. Let
![]() (71)
(71)
denote the characteristic polynomial of the two by two matrix A in (7). Note, that if ![]() and
and ![]() and
and ![]() then
then ![]() and
and![]() . Here
. Here
![]() (72)
(72)
![]() (73)
(73)
Define
![]() (74)
(74)
So if ![]() then we have the polynomial
then we have the polynomial
![]() (75)
(75)
If this polynomial is a Routh Hurwitz polynomial, i. e. the roots lie in![]() , then the roots of
, then the roots of
![]() (76)
(76)
lie in the interior ![]() of the unit circle. Now compute
of the unit circle. Now compute
![]() (77)
(77)
and
![]() (78)
(78)
Also
![]() (79)
(79)
If
![]() (80)
(80)
and ![]() the fixed point of S is stable, then by the Routh Hurwitz criterion
the fixed point of S is stable, then by the Routh Hurwitz criterion
![]() (81)
(81)
![]() (82)
(82)
But this implies, by adding these two inequalities, that
![]() (83)
(83)
However, then
![]() (84)
(84)
A contradiction to (80). So if ![]() is stable, then
is stable, then![]() . Assume now that
. Assume now that![]() . If
. If ![]() is stable, then
is stable, then
![]() (85)
(85)
by the Routh Hurwitz criterion. We shall find the fixed points of S, with![]() . From the second coordinate find
. From the second coordinate find
![]() (86)
(86)
Then the first coordinate gives
![]() (87)
(87)
But the denominator is positive if ![]() is stable, by the above. Hence the treatment benefits the patient, because we assume that
is stable, by the above. Hence the treatment benefits the patient, because we assume that![]() , and
, and
![]() (88)
(88)
and we are lowering ![]() by
by![]() .
.
Now suppose
![]() (89)
(89)
The assumption ![]() implies that -1 is a root of
implies that -1 is a root of
![]()
Since![]() , we have
, we have
![]() (90)
(90)
We claim that ![]() is stable, when
is stable, when![]() . But then
. But then ![]() and
and ![]() are the distinct eigenvalues of A. So there is a change of basis matrix D such that
are the distinct eigenvalues of A. So there is a change of basis matrix D such that
![]() (91)
(91)
Clearly both
![]() (92)
(92)
and
![]() (93)
(93)
have unique fixed points ![]() and
and ![]() and we clearly have
and we clearly have
![]() (94)
(94)
We need the following definition.
Definition. A fixed point ![]() of
of ![]() is stable if given an open neighbourhood
is stable if given an open neighbourhood ![]() of
of ![]() there exists an open neighbourhood
there exists an open neighbourhood ![]() of
of ![]() such that for all
such that for all ![]()
![]() (95)
(95)
for all![]() . A fixed point is unstable if it is not stable.
. A fixed point is unstable if it is not stable.
Now observe, that
![]() (96)
(96)
![]() (97)
(97)
Notice that
![]() (98)
(98)
in the max norm
![]() (99)
(99)
because
![]() (100)
(100)
But now stability follows from the estimate
![]() (101)
(101)
and this implies that ![]() is stable, because S and
is stable, because S and ![]() are conjugate:
are conjugate:
![]() (102)
(102)
If![]() , then we get the estimate when
, then we get the estimate when ![]()
![]() (103)
(103)
as![]() . So
. So ![]() is unstable and since
is unstable and since ![]() and S are conjugate,
and S are conjugate, ![]() is unstable.
is unstable.
3. Models of Cancer Growth
Consider the mapping
![]() (104)
(104)
where
![]() (105)
(105)
and
![]() (106)
(106)
The matrix here is denoted A. T maps ![]() to itself. g is a vector of birth rates and
to itself. g is a vector of birth rates and![]() . The
. The![]() . Finally
. Finally ![]() . Also put
. Also put
![]() (107)
(107)
![]() (108)
(108)
![]() (109)
(109)
![]() (110)
(110)
![]() (111)
(111)
![]() (112)
(112)
C is cancer ![]() are growth factors and
are growth factors and ![]() are growth inhibitors,
are growth inhibitors,![]() .
.
Proposition 1 The characteristic polynomial ![]() of A is
of A is
![]() (113)
(113)
![]() (114)
(114)
Proof. With![]() ,
,
![]() (115)
(115)
![]() (116)
(116)
Decompose ![]() after the last column to obtain, assuming the formula for
after the last column to obtain, assuming the formula for ![]() holds
holds
![]() (117)
(117)
![]() (118)
(118)
![]() (119)
(119)
![]() (120)
(120)
![]() (121)
(121)
![]() (122)
(122)
![]() (123)
(123)
Suppose henceforth, that
![]() (124)
(124)
Then the characteristic polynomial of A is
![]() (125)
(125)
So the eigenvalues are ![]() and since
and since
![]() (126)
(126)
is a factor of![]() , then
, then
![]() (127)
(127)
are eigenvalues of A, where
![]() (128)
(128)
![]() (129)
(129)
For the moment assume![]() . Define the matrix of eigenvectors of A by
. Define the matrix of eigenvectors of A by
![]() (130)
(130)
We shall find formulas for the complements
![]() (131)
(131)
of D and the determinant of D,![]() .
.
Proposition 2 For ![]() we have
we have
![]() (132)
(132)
For ![]()
![]() (133)
(133)
Proof. Suppose![]() . We are deleting row r. So in column
. We are deleting row r. So in column ![]() there is only one nonzero element
there is only one nonzero element![]() . Decomposing after this column and then after row one we get a matrix with zeroes under the diagonal. The signs here are
. Decomposing after this column and then after row one we get a matrix with zeroes under the diagonal. The signs here are
![]() (134)
(134)
![]() is the sign on the complement
is the sign on the complement ![]() and
and ![]() is the sign on the complement to
is the sign on the complement to![]() . It is in row
. It is in row ![]() and column
and column ![]() and we delete two rows.
and we delete two rows. ![]() is the sign on
is the sign on![]() . Hence
. Hence
![]() (135)
(135)
which is what we wanted to prove.
Now suppose that![]() . For
. For ![]() we get a matrix with zeroes under the diagonal, so
we get a matrix with zeroes under the diagonal, so
![]() (136)
(136)
Now consider the case![]() . Write
. Write![]() . After decomposing after rows
. After decomposing after rows ![]() and row one column two in
and row one column two in ![]() we are left with
we are left with
![]() (137)
(137)
Decompose after rows![]() , to get
, to get
![]() (138)
(138)
which gives
![]() (139)
(139)
The proposition follows, because
![]() (140)
(140)
Proposition 3
![]() (141)
(141)
Proof. We have
![]() (142)
(142)
Initially let![]() . Now
. Now
![]() (143)
(143)
when ![]() and
and
![]() (144)
(144)
when![]() . Now decompose after the last column to get
. Now decompose after the last column to get
![]() (145)
(145)
If
![]() (146)
(146)
(![]() in the statement of the proposition) we get
in the statement of the proposition) we get
![]() (147)
(147)
Now we shall use induction over q to prove the formula in the statement of the proposition. Decompose after the last row
![]() (148)
(148)
![]() (149)
(149)
![]() (150)
(150)
![]() (151)
(151)
In B we have decomposed after ![]() and then after the rows
and then after the rows ![]() and in the remaining matrix decomposed after row p and column
and in the remaining matrix decomposed after row p and column![]() . The signs here are
. The signs here are
![]() (152)
(152)
The proposition follows.
The aim of our computations is to show that there exists an affine vector field X on ![]() such that the time one map is
such that the time one map is
![]() (153)
(153)
Let ![]() denote an integral curve of X through
denote an integral curve of X through![]() . Then we shall find a formula for
. Then we shall find a formula for
![]() (154)
(154)
First notice that
![]() (155)
(155)
if
![]() (156)
(156)
and
![]() (157)
(157)
which we assume. We have used that
![]() (158)
(158)
So the eigenvectors in D are linearly independent, hence
![]() (159)
(159)
Now define when ![]()
![]() (160)
(160)
where![]() . The flow of Y is
. The flow of Y is
![]() (161)
(161)
![]() (162)
(162)
where we denote the last vector![]() . This is readily shown by differentiating
. This is readily shown by differentiating ![]() with respect to t
with respect to t
![]() (163)
(163)
![]() (164)
(164)
![]() (165)
(165)
Now we get
![]() (166)
(166)
![]() (167)
(167)
![]() (168)
(168)
It follows that ![]() is the flow of Y. Now require
is the flow of Y. Now require
![]() (169)
(169)
that is
![]() (170)
(170)
We shall require
![]() (171)
(171)
because then the time one map of Y is
![]() (172)
(172)
Now define
![]() (173)
(173)
Then the flows of X and Y are related by
![]() (174)
(174)
But then the time one map of X is
![]() (175)
(175)
which is what we wanted.
Theorem 4 Assume, that
![]() (176)
(176)
and
![]() (177)
(177)
We have the formula
![]() (178)
(178)
![]() (179)
(179)
![]() (180)
(180)
![]() (181)
(181)
Proof. We use the formula
![]() (182)
(182)
![]() (183)
(183)
![]() (184)
(184)
We have
![]() (185)
(185)
and then
![]() (186)
(186)
for ![]() and for
and for ![]()
![]() (187)
(187)
We shall write
![]() (188)
(188)
and then we have
![]() (189)
(189)
When ![]() then
then
![]() (190)
(190)
while for ![]() we have
we have
![]() (191)
(191)
Notice that
![]() (192)
(192)
Continuing from (184)
![]() (193)
(193)
![]() (194)
(194)
![]() (195)
(195)
![]() (196)
(196)
![]() (197)
(197)
So this gives the first term in
![]() (198)
(198)
Note that
![]() (199)
(199)
Now we have
![]() (200)
(200)
![]() (201)
(201)
![]() (202)
(202)
Hence the ![]() contribution is from (184)
contribution is from (184)
![]() (203)
(203)
![]() (204)
(204)
and the ![]() contribution is
contribution is
![]() (205)
(205)
So
![]() (206)
(206)
![]() (207)
(207)
![]() (208)
(208)
![]() (209)
(209)
The theorem follows.
Now suppose that![]() . Then
. Then
![]() (210)
(210)
![]() . We shall require that
. We shall require that![]() . Now define the matrix
. Now define the matrix
![]() (211)
(211)
The first column is denoted![]() , the second is denoted
, the second is denoted![]() . Here
. Here![]() , both in
, both in![]() . Notice that
. Notice that
![]() (212)
(212)
![]() (213)
(213)
![]() (214)
(214)
![]() (215)
(215)
Now as in [1] we get
![]() (216)
(216)
![]() (217)
(217)
![]() (218)
(218)
![]() (219)
(219)
and similarly
![]() (220)
(220)
![]() (221)
(221)
![]() (222)
(222)
So
![]() (223)
(223)
because we assume that![]() . Exactly as before we get
. Exactly as before we get
Proposition 5 For ![]()
![]() (224)
(224)
and for ![]()
![]() (225)
(225)
Also
![]() (226)
(226)
Finally
![]() (227)
(227)
since
![]() (228)
(228)
Proof. The proposition follows immediately from proposition 2 and 3.
The flow of
![]() (229)
(229)
is, for ![]()
![]() (230)
(230)
We want to have that this equals for![]() , the matrix
, the matrix
![]() (231)
(231)
Thus
![]() (232)
(232)
![]() (233)
(233)
Remember the formula
![]() (234)
(234)
Define when![]() .
.
![]() (235)
(235)
where![]() . The flow of Y is
. The flow of Y is
![]() (236)
(236)
![]() (237)
(237)
where the last vector is denoted![]() . To see this compute
. To see this compute
![]() (238)
(238)
![]() (239)
(239)
Now we also get
![]() (240)
(240)
![]() (241)
(241)
So ![]() is the flow of Y. We need to have
is the flow of Y. We need to have
![]() (242)
(242)
because then the time one map of Y is
![]() (243)
(243)
Then define
![]() (244)
(244)
The flows are related by
![]() (245)
(245)
But then the time one map of X is
![]() (246)
(246)
which is what we intended to find.
Theorem 6 When ![]() then
then
![]() (247)
(247)
![]() (248)
(248)
![]() (249)
(249)
Proof. We have the following computation
![]() (250)
(250)
![]() (251)
(251)
And we want to have
![]() (252)
(252)
![]() (253)
(253)
that is
![]() (254)
(254)
But we have arranged that
![]() (255)
(255)
so we get
![]() (256)
(256)
![]() (257)
(257)
Denote the two by two matrix in the last line
![]() (258)
(258)
We can also compute the first term in
![]() (259)
(259)
omitting the factor ![]()
![]() (260)
(260)
![]() (261)
(261)
![]() (262)
(262)
![]() (263)
(263)
hence the first term in
![]() (264)
(264)
Now
![]() (265)
(265)
![]() (266)
(266)
The ![]() contribution is omitting the factor
contribution is omitting the factor ![]()
![]() (267)
(267)
![]() (268)
(268)
![]() (269)
(269)
![]() (270)
(270)
The ![]() contribution is, omitting the factor
contribution is, omitting the factor ![]()
![]() (271)
(271)
![]() (272)
(272)
![]() (273)
(273)
![]() (274)
(274)
where
![]() (275)
(275)
The theorem follows.
Now assume that![]() . Define
. Define
![]() (276)
(276)
Proposition 7 For q odd and ![]()
![]() (277)
(277)
and for ![]() and q odd
and q odd
![]() (278)
(278)
For q even and ![]()
![]() (279)
(279)
and for ![]() and q even
and q even
![]() (280)
(280)
Also
![]() (281)
(281)
when q is odd and
![]() (282)
(282)
when q is even.
Proof. (277) q odd. We are deleting the row r with![]() . Decompose after that column with
. Decompose after that column with ![]() in it and row one column two. The sign on
in it and row one column two. The sign on ![]() is
is
![]() (283)
(283)
The first sign here is the sign when decomposing after row![]() , except the row with
, except the row with![]() . The second sign is the sign on the complement to
. The second sign is the sign on the complement to![]() . The third sign is the sign on
. The third sign is the sign on![]() . The last sign is the sign on
. The last sign is the sign on ![]() in column r and row one. (277) follows. Now let
in column r and row one. (277) follows. Now let![]() . Write
. Write![]() . Decomposing after rows
. Decomposing after rows ![]() to give
to give
![]() (284)
(284)
We have the sign
![]() (285)
(285)
on![]() . And we have the sign
. And we have the sign
![]() (286)
(286)
on column ![]() and row one. Hence the formula.
and row one. Hence the formula. ![]() is obvious. And the formulas for q even follow similarly. (281) q odd. First let
is obvious. And the formulas for q even follow similarly. (281) q odd. First let ![]() and
and![]() . Then
. Then
![]() (287)
(287)
For ![]() we get
we get
![]() (288)
(288)
We have, decomposing after the last column
![]() (289)
(289)
![]() (290)
(290)
![]() (291)
(291)
![]() (292)
(292)
Here
![]() (293)
(293)
For q even we get
![]() (294)
(294)
Now decompose after the last column
![]() (295)
(295)
Now we get decomposing after the last column
![]() (296)
(296)
![]() (297)
(297)
![]() (298)
(298)
![]() (299)
(299)
where
![]() (300)
(300)
In the determinant B, we have decomposed after row 2 to![]() . In the remaining determinant decompose after row one and column
. In the remaining determinant decompose after row one and column![]() . The proposition follows.
. The proposition follows.
Define for ![]() and
and ![]()
![]() (301)
(301)
From Proposition 7, we get
Proposition 8 For q odd and ![]()
![]() (302)
(302)
and for q odd and ![]()
![]() (303)
(303)
For q even and ![]()
![]() (304)
(304)
and for q even and ![]()
![]() (305)
(305)
Also for q odd and ![]()
![]() (306)
(306)
and for q odd and ![]()
![]() (307)
(307)
For q even and ![]()
![]() (308)
(308)
and for q even and ![]()
![]() (309)
(309)
Finally for q odd
![]() (310)
(310)
and
![]() (311)
(311)
for q even.
4. An ODE Model
In [1] we also considered a three dimensional ODE model of cancer growth in the variables ![]() cancer, growth factors and growth inhibitors, respectively. Analogous to what we did in section three define a mass action kinetic system
cancer, growth factors and growth inhibitors, respectively. Analogous to what we did in section three define a mass action kinetic system
![]() (312)
(312)
![]() (313)
(313)
![]() (314)
(314)
![]() (315)
(315)
![]() (316)
(316)
![]() (317)
(317)
Here the complexes are![]() ,
, ![]() ,
, ![]() ,
, ![]() ,
, ![]() ,
, ![]() This defines the rate constants. For a reaction
This defines the rate constants. For a reaction
![]() (318)
(318)
the forward reaction rate is denoted ![]() and the reverse reaction rate is denoted
and the reverse reaction rate is denoted![]() . The differential equations are
. The differential equations are
![]() (319)
(319)
![]() (320)
(320)
![]() (321)
(321)
We shall find a polynomial giving candidates of singular points of this vector field.
From![]() , we find
, we find
![]() (322)
(322)
where![]() . From
. From![]() , we find
, we find
![]() (323)
(323)
for![]() . Inserted into
. Inserted into ![]() we get
we get
![]() (324)
(324)
We can then multiply with
![]() (325)
(325)
and define the constants
![]() (326)
(326)
to obtain the polynomial of degree ![]()
![]() (327)
(327)
![]() (328)
(328)
if we assume that![]() . There is a relation between the ODE model of this chapter, with vector field
. There is a relation between the ODE model of this chapter, with vector field
![]() (329)
(329)
and the discrete dynamical system of section three, see also [1] . Linearize the vector field at a singular point ![]() and set
and set
![]() (330)
(330)
Also define the Euler map
![]() (331)
(331)
for![]() . This is an approximation to the flow of h. If we let
. This is an approximation to the flow of h. If we let
![]() (332)
(332)
![]() (333)
(333)
![]() (334)
(334)
![]() (335)
(335)
![]() (336)
(336)
![]() (337)
(337)
and
![]() (338)
(338)
![]() (339)
(339)
then you obtain a discrete model T of section three.
Example Let ![]() and define the rate constants
and define the rate constants ![]() and
and![]() ,
,![]() . Then there are two positive singular points.
. Then there are two positive singular points.
5. Summary
In this paper, we considered a discrete mathematical model and an ODE model of cancer growth in the variables ![]() cancer, growth factors and growth inhibitors, respectively. We have shown that this model is a threshold model. If
cancer, growth factors and growth inhibitors, respectively. We have shown that this model is a threshold model. If ![]() and
and
![]() (340)
(340)
then cancer grows, and if the reverse inequality holds, cancer is eliminated. We also proposed personalized treatment using the simple model of cancer growth in the introduction and the ODE model of section four.