Assessing Genetic Variation and Relationships among a Mini Core Germplasm of Sesame (Sesamum indicum L.) Using Biochemical and RAPD Markers ()
1. Introduction
Sesame (Sesamum indicum L.) is a warm weather crop, often growing under stress conditions. Its seed contains 20% - 25% protein and 50% - 60% oil. Sesame oil has excellent stability due to the presence of natural antioxidants such as sesamolin, sesamin, and sesamol [1] . Myanmar, India and China are the most important sesame producers followed by Ethiopia, Nigeria, Sudan, Uganda and Tanzania in Africa [2] . In Benin, sesame is grown in the north-western and north-eastern zones, as a subsistence crop either for its leaves or seeds, which are roasted for food. It is anticipated that commercial sesame cultivation will increase in this country. The elite cultivar 38-1-7 was introduced to most major sesame planting areas in Benin over the last 20 years according to farmers. The decreasing crop yield according to farmers can be attributed to its cultivation in un-irrigated areas, lack of varietal replacement through development of hybrids. According to Pathak et al. [3] low productivity has been partially attributed to a lack of improved varieties tolerant to biotic and abiotic stresses like diseases, pests and drought. The low seed yield of sesame is a consequence of a lack in breeding attention. Sesame production is also limited by lack of uniform maturity of capsules, and seed shattering [4] .
The main challenges associated with sesame cultivation are drought and heat stress at certain periods of the growing season [5] . Though variations in climatic and edaphic conditions affect sesame yields and performance, the major constraints identified in growing sesame in most countries are instability in yield, lack of wider adaptability, drought, nonsynchronous maturity, poor stand establishment, lack of response to fertilizer application, profuse branching, lack of seed retention, low harvest index and susceptibility to insect pests and pathogens [6] . Furthermore, global change is predicted to increase the severity of drought and heat stress in warm temperate to tropical areas with potentially large effects on yield [7] . Thus, building on genetic diversity to find suitable varieties and breeding material for stressed conditions is of top priority to continue sesame cultivation in traditional areas. Thousands of sesame accessions have been collected, and a lot of work on morpho-agronomic characterisation has been done [8] [9] . A very wide diversity is available in existing germplasms and desirable traits have been reported in those germplasms [10] . Some agronomic traits, useful for yield improvement, have been identified, e.g. non shattering capsule, long capsule, pests and diseases resistance. However, little information on the molecular and seed esterase variation is available for S. indicum in Benin; and also how accessions from Benin relate to other accessions from the world.
Genetic diversity of crops is important for sustainable development and food security. It is also important for selection of parents that can be used in plant breeding programs. Improvement of a crop species requires good knowledge on genetic diversity. Pathak et al. [3] reviewed sesame diversity studies carried out using various methods such as morphological, biochemical and molecular markers.
Moreover, seed storage proteins [11] and isozymes [12] [13] have proven to be reliable markers for assessing diversity in sesame, since they do not change with environmental conditions. Several types of molecular markers have been developed and applied to sesame genotyping, such as random amplified polymorphic DNA (RAPD) [14] [15] , simple sequence repeats (SSR) [16] , inter-simple sequence repeats (ISSR) [17] , amplified fragment length polymorphism (AFLP) [18] , sequence-related amplified polymorphisms (SRAP) [19] and expressed sequence tags-SSR (EST-SSR).
In this study, we analysed the genetic diversity of a mini core sample of morphologically diverse accessions using biochemical and molecular markers, and assessed the degree of congruence between the agro-morphological biochemical, and molecular markers in Sesamum indicium.
2. Materials and Methods
2.1. Plant Material
A total of 15 accessions of S. indicum were included in the investigation. 10 were obtained from core collections established at CAAS, Wuhan (China); one was obtained from INRAB, Niaouli (Benin), and the rest from the demonstration garden at Huajiachi Campus, Zhejiang University. Characteristics are provided in Table 1.
2.2. DNA Extraction
About 15 seeds per accessions were used for extraction. The seeds were ground with a pestle and mortar. The powder was thoroughly mixed and homogenised in 600 μl of extraction buffer (containing 2% CTAB, 100 mMTris-HCl pH 8.0, 20 mM EDTA pH 8.0, 1.4 M NaCl, 2% ME) in a 1.5 microcentrifuge tube. The homogenate was maintained for 40 min at 55˚C in water bath. The tubes were
![]()
Table 1. Characteristics of the 15 accessions investigated in this study.
NB: x means unknown origin; -Mean values and standard error (in parentheses) for the seed parameters.
kept for 10 min to allow them to return to room temperature. An equal volume of chloroform/isoamyl alcohol (24:1) was added to the tubes and they were centrifuged at 13,000 rpm at 15˚C for 15 min. The supernatant was transferred to a clean tube. 1/10 volume of 10% CTAB, 1/3 volume of 70% alcohol and 2/3 volume of chloroform/isoamyl alcohol were added to the tubes, and they were centrifuged at 13,000 rpm at 15˚C for 15 min. The supernatant was transferred to a fresh tube and DNA precipitated by adding an equal volume of precipitation solution (1% CTAB, 50 mMTris-HCl pH 8.0, 10 mM EDTA pH 8.0, 1% ME). The tubes were kept static for more than 30 min and then centrifuged at 5500 rpm at 4˚C for 10 min. The tubes were allowed to drain. The DNA precipitate was dissolved in 300 μl TE (10 mMTris-HCl pH 8.0, 1 mM EDTA pH 8.0, 1M NaCl, 1.5% SDS, 1 μg/ml RNase) and incubated for a minimum of 60 min at 37˚C in water bath. An equal volume of solution of phenol/chloroform, prepared in a ratio of 1:1, was added to the tube and mixed gently and the tube was centrifuged again. DNA was precipitated using isopropanol. The DNA pellet was washed with 70% ethanol and absolute ethanol, and air-dried at room temperature for 3 hours. Final pellet was solubilized in 200 μl TE and kept at −20˚C.
2.3. RAPD Amplification
Oligonucleotide primers of 10-mer from the series OPS (Operon Technologies/Shanghai Shending) were tested for their ability to amplify scorable and reproducible fragments.
The polymerase chain reactions (PCRs) were performed following Williams et al. [20] with some modifications. Each 15 μl PCR reaction mixture consisted of approximately 10 ng template DNA; 400 μg primer; 1.0 U Taq DNA polymerase; 200 μmoldNTP; 2.0 mmol Mg2+. The PCR reaction was carried out in a DNA- Thermal-Cycler 480 PCR under the following programmed conditions: an initial denaturing step of 5 min at 94˚C, 40 cycles consisting each of 1 min at 94˚C, 1 min 38˚C, and 2 min at 72˚C with a final elongation step of 10 min at 72˚C.
The products of the reaction were electrophoresed in 1xTAE buffer on 1.4% agarose gels containing 0.2 μg/μl of ethidium bromide for about 3 hours. The gels were photographed under UV light with Polaroid film.
2.4. Seed Storage Proteins
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) of seed storage protein was carried out using a discontinous gel technique as described by ISTA [21] rules 8.6.A.3. Five seeds per accession of sesame were crushed with 150 μl extraction buffer in 1.5 ml centrifuge tubes.
2.5. Esterase
Three seeds (or seedlings) per accession of sesame were grinded (or macerated) with 100 μl buffer (Tris-Hcl and saccharose). Samples were centrifuged for 5 min at 10,000 tr/min in an Eppendorf microcentrifuge. The supernatant (25 μl) was electrophoresed on polyacrylamide of 2.5% (pH 6.7) in the concentration and 8.75% (pH 8.9) in the separation gel. The electrophoretic separation was initiated with 150 V, and voltage was slowly raised to 300 V after the tagging dye had passed to the separation gel. Electrophoresis was carried out at 10˚C - 20˚C for approximately 2 - 3 hr. The gels were stained in solution containing 20 mg α-naphtyl acetate, 20 mg β-naphtyl acetate and 150 mg Fast blue RR salt in 200 ml 0.1 M sodium phosphate solution. Gels were incubated at 37˚C until bands were visualized.
2.6. Data Analysis
The RAPD assays were performed in triplicate and only those patterns obtained at least twice were scored. Fragments were manually scored as presence (1) or absence (0) of amplification product for each primer and used to calculate Dice’s coefficient of similarity for each pair of accessions as follows:
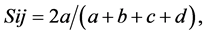
equivalent to the coefficient of Nei & Li [22] where Sij is the similarity between two individuals, i and j; a is the number of bands present in both i and j; b is the number of bands present in i and absent in j; c is the number of bands present in j and absent in i and d is the number of bands absent in both. The similarity value found were used to group accessions via Unweighted Pair Group Method (UPGMA). NTSYSpc version 2.2 [23] was used to perform all analyses.
The banding patterns and schematic diagrams (zymograms) were recorded. The relative migration (Rf values) of each band was measured in each zymogram for every accession studied using the following equation: Rf Value = Distance migrated by the enzyme band from the cathodal edge/Distance migrated by the dye marker. Data were recorded as the presence or absence and relative band intensity. For genetic diversity relationships, cluster analysis was performed using NTSYS-pc version 2.0.
3. Results and Discussion
3.1. Plant Material Analysis
One of the most important attributes for introducing sesame grains in the market is seed color. Seed coat color is an important agronomic trait in sesame, as it is associated with seed biochemical properties, antioxidant content and activity and even disease resistance of sesame. There is a wide variability in sesame seed coat color (Table 1), which varies from white to black. Due to the importance of this trait for the export market, seed color is a central target in sesame breeding programs [24] [25] .
3.2. RAPD Analysis
3.2.1. DNA Template
The concentrations of template DNA tested were 2.3, 4.6, 9.2, 18.4, 36.8 ng.
Approximately 10 ng of DNA template gave maximum number of reproducible bands and thus was used in this experiment. These results showed that DNA extraction from seed, without germinating them, is applicable in sesame. Generally, there is no dormancy in sesame. However, accession ZZM2431 showed short-term seed dormancy. Ashri [26] also reported six months seed dormancy in a Mexican cultivar, but that dormancy could be broken by soaking seed in GA3 solutions. Therefore, using seed for DNA extraction could escape dormancy observed in sesame and germination efforts.
3.2.2. Fingerprints Patterns and Primer Selection
RAPD markers were used to evaluate the extent of genetic diversity among sesame accessions. As molecular markers, it is comparatively cheap and simple [27] [28] . Single arbitrary 10-mer primers from the series OPS were tested for their ability to amplify scorable and reproducible DNA fragments. Primers resulting in complex amplification pattern or unreproducible PCR products per accession were excluded for the statistical analysis. Six primers (Table 2) yielding consistent and distinct amplification products were then included in this analysis.
3.2.3. Genetic Variation
A total of 53 fragments were produced by the 6 primers across the 15 materials. The number of fragment per primer varied from 4 to 14 (Table 2), with an average of 8.83. Between 2 and 13 polymorphic bands were scored for each primer. Out of 53 generated fragments, 48 (90.6%) were polymorphic, indicating that there was considerable variation at DNA level in the sesame germplasm investigated.
3.2.4. Accession-Specific Markers
Twelve accession-specific RAPD markers were observed; these fragments were unique to six different accessions. The five RAPD bands OPS29-b, OPS29-l, OPS32-b, OPS32-e and OPS38-a were unique to the single accession “Balinzhima”, and the bands OPS01-c, OPS29-c and OPS35-a were unique to accession ZZM0611. The fragments OPS01-b and OPS01-f were only present in ZZM2918 and INRAB38-1-7 respectively. Similarly ZZM2874 and “Laohongzhulian” were sufficiently distinguishable from all the others by RAPD bands OPS29-a and OPS01-n respectively (Figures 1-5). This discrimination was expected as the
![]()
Table 2. RAPD primers and number of fragment scored for each primer.
![]()
Figure 1. PCR-RAPD profile of 15 accessions of sesame generated by primer OPS1. (Name of genotypes 1 to 15 is given in Table 1).
![]()
Figure 2. PCR-RAPD profile of 15 accessions of sesame generated by primer OPS 22. (Name of genotypes 1 to 15 is given in Table 1).
![]()
Figure 3. PCR-RAPD profile of 15 accessions of sesame generated by primer OPS 29. (Name of genotypes 1 to 15 is given in Table 1).
![]()
Figure 4. PCR-RAPD profile of 15 accessions of sesame generated by primer OPS 32. (Name of genotypes 1 to 15 is given in Table 1).
![]()
Figure 5. PCR-RAPD profile of 15 accessions of sesame generated by primer OPS 38. (Name of genotypes 1 to 15 is given in Table 1).
objective to find out diversity in minute molecular level and can be useful for obtaining cultivar/genotype specific profiles as they showed high level of polymorphism among the selected accessions. These unique RAPD markers found in our study could be used to identify selected sesame accessions. In other studies, Sarkar et al. [14] found height primers resulted unique bands. Primer OPA-2, OPA-4, OPA-5, OPA-9, OPA-18, OPM-10, OPM-16, OPM-20 were able to reveal > 60% polymorphism among the accessions selected.
3.2.5. Genetic similarity
Pair-wise similarity coefficient among the 15 accessions ranged from 0.343 to 0.897 (Table 3). The pair of accessions ZZM0795 and Balinzhima had the lowest similarity coefficient of 0.343, while ZZM0829 and ZZM3186 were the most distant. Out of 105 pair of sesame combinations, 16 had a similarity coefficient of less than 0.50, indicating a wide genetic difference between them. A very high level of polymorphism has been observed with the of RAPD markers indicating a wide and diverse genetic base of the sesame germplasm analysed by Bath et al. [27] . However, in 38 pairs of sesame combinations, the similarity coefficient was greater than 0.70, such as accessions 38-1-7 (from Benin, West-Africa) and ZZM2737 (from Zhejiang, China) showing great similarity of 0.878. These results revealed that most of the accessions showed high degree of genetic similarity at the molecular level following RAPD analysis. Bhat et al. [27] , in their stu- dy, also observed association between variation among sesame collections and their geographic origin.
3.2.6. Cluster Analysis
Based on Dice coefficient similarity matrix using UPGMA, the 15 accessions were grouped into two clusters I and II at the point of 0.64 genetic similarity, and one independent accession, “Balinzhima” at the point 0.43 (Figure 6).
![]()
Table 3. Pair-wise similarity of the 15 accessions.
(Name of genotypes 1 to 15 is given in Table 1).
![]()
Figure 6. Dendrogram generated for 15 sesame accessions from 48 polymorphic RAPD markers by cluster (UPGMA) analysis.
Divergent genotypes may have good breeding value. In cluster I, there were
five accessions, of which ZZM0611 was the most divergent. Cluster II which con- tained the other nine sesame accessions, were further divided into two sub-clusters at the point of 0.70 similarity. Although the cluster analysis classified accessions into several clusters or sub-clusters, the accessions in each group could not be associated with their locality of origin or seed coat colour. Outcrossing in sesame can reach 68% [26] . Also several accessions were acquired as seed and the origin can thus not be guaranteed. All these observations are supported by evidences from the investigation of Zhao et al. [12] . Out of the 5 isozymes studied in 18 sesame accessions, only esterase showed diversity between accessions and within an accession. Such a variation within an individual accession led to conclude that sesame accessions investigated were all heterogeneous. The objective of our experiment was prevalence of gross genetic differences among accessions. In such conditions, about 15 seeds per accession were bulked for DNA extraction, and thus the frequency of RAPD profiles per accession was probably hided.
The primers individually could group the 15 accessions into 3 or 4 groups each. However, none of the individual primers could not cluster these germ- plasms into region-specific or seed colour-specific groups. Our results were in line with recent works on sesame diversity using RAPD markers [14] [15] . This lack of strong association between geographical distribution and classification based on molecular markers in sesame has been observed also in other studies [29] [30] , and may reflect exchange of sesame germplasm among widely separated locations [30] . Our results support the previous studies wherein a very low relationship between geographic origin and genetic diversity has been reported in sesame germplasm based on morpho-agronomic traits [31] , that may be due to exchange over short distances and micro-environment influences. Nevertheless, Pham [30] found a relatively high correlation (r = 0.88, P < 0.001) between the agro-morpho- logical and RAPD data in terms of relationship between sesame accessions.
3.3. Seed Protein and Esterase Analysis
The diversity estimates based on esterase analysis were not in consensus with RAPD results this might be due to limitations of biochemical markers for resolving intra species differences. Resolution of individual bands was best in seed-EST, seedling-EST and seed storage protein in descending order.
3.3.1. Seed Storage Protein
The SDS-polyacrylamide gel electrophoregram of seed storage protein of the 15 accessions of sesame investigated is shown in Figure 7 and Figure 8. The greatest number of banding patterns was recognized in seed storage protein. But, out of 31 bands 20 were monomorphic and out of the remaining 11 polymorphic bands, 8 were similar. Cluster analysis based on SDS-PAGE analysis of protein peptides were more reliable where most of the accessions of each group were collected from the same geographic location. There are great similarities in the overall polypeptide profiles of seed protein from the accessions, indicating a common origin for these sesame germplasms. This finding is supported by Fazal et al. [32] who reported a limited level of intra specific variation for seed protein among sesame germplasm. However, SDS-PAGE techniques are successfully used for studying the genetic diversity in several researches [33] [34] . Based on presence or absence of some bands, it was possible to separate accessions into groups. But this high similarity between protein patterns of the 15 accessions makes seed storage protein unsuitable markers for this study.
3.3.2. Seed Esterase
The zymogram for esterase extracted from sesame seed is given in Figure 9
![]()
Figure 7. Electropherogram SDS-PAGE of seed proteins of 15 accessions of sesame (Name of genotypes 1 to 15 is given in Table 1).
![]()
Figure 8. Diagrammatic representation of the electrophoregram SDS-PAGE of 15 accessions of sesame (Name of genotypes 1 to 15 is given in Table 1).
![]()
Figure 9. Zymogram of seed-esterase of 15 accessions of sesame (Name of genotypes 1 to 15 is given in Table 1).
and Figure 10. Seven of the 14 bands were monomorphic. The black and grey coat seed accessions namely ZZM2812, ZZM2737, Laohongzhulian, Jiaoganzhu and Balinzhima lacked band at 0.39 mobility. This absence of band at Rf of 0.39 may probably be associated with good storage potential [35] . The genetic similarity, based on presence/absence of bands, varied from 0.71 to a maximum of 1.00 indicating high level of prevalent similarity. The white seed coat accession ZZM0611 was the most diverse among the 15 accessions investigated. In addition, another analysis based on presence/absence and staining intensity of bands showed more diversity among the 15 sesame accessions. The dissimilarity coefficient varied from 0.0 to a maximum of 1.1 (Figure 11). Since accessions differing in seed colour had identical esterase patterns, and vice versa, accessions with similar seed colour had different esterase patterns, there is most likely no correlation between esterase banding patterns and seed coat colour. Similarly, similarity of seed-esterase patterns was not correlated with geographic origin.
3.3.3. Seedling Esterase
The zymogram for esterase extracted from sesame seedling is given in Figure 12 and Figure 13. Nine of the 13 bands were monomorphic. The accession 38-1-7
![]()
Figure 10. Diagrammatic representation of the zymogram of seed esterase of 15 accessions of sesame (Name of genotypes 1 to 15 is given in Table 1).
![]()
Figure 11. Dendrogram of 15 accessions based on data generated from seed-esterase zymogram.
![]()
Figure 12. Zymogram of seedling-esterase of 15 accessions of sesame (Name of genotypes 1 to 15 is given in Table 1).
![]()
Figure 13. Diagrammatic representation of the zymogram of seedling-esterase of 15 accessions of sesame (Name of genotypes 1 to 15 is given in Table 1).
is distinct from all the other accessions as shown in the cluster analysis (Figure 14). Since accessions differing in seed colour had identical esterase patterns, and vice versa, accessions with similar seed colour had different esterase patterns, there is most likely no correlation between esterase banding patterns and seed coat colour. However, all accessions from China were grouped together. Similarity in seedling-esterase patterns was correlated with geographic origin, China and West-Africa.
Use of biochemical methods eliminates the environmental influence, reveal differences between seed storage proteins or enzymes encoded by different alleles at one (allozymes) or more gene loci (isozymes). The usefulness is limited due to their inability to detect low levels of variation. Thus the major advantage of DNA-based techniques being that they analysed the variation at DNA level
![]()
Figure 14. Dendrogram of 15 accessions based on combination of data generated from seed- and seedling-esterase zymograms. (absence: 0 or presence: 1/intensity of band).
itself, excluding all environmental influences. For example, Microsatellites and SSRs are also considered a powerful tool to investigate plant variability [36] . But, it has been assumed that in plant breeding, diversity can be reduced using biochemical molecular techniques. Present study was carried out on diversity of fifteen sesame accessions collected from different parts of the world. The examination of S. indicum germplasm accessions based on biochemical markers resulted in moderate differences between accessions. However, Zhao et al. [12] and Nanthakumar et al. [13] reported large variability among sesame accessions using esterase isozyme. Likewise, Leelambike et al. [37] used esterase to confirm diversity among Mucuna species. By using qualitative differences (presence or absence of some bands) and also quantitative differences (relative intensity of bands) of seed-esterase bands, most of the accessions investigated in this study could be differentiated. But the results obtained by clusters did not form robust groups, which could be associated with geographic origin or seed coat colour. According to Laurentin and Karlovsky [38] , the traditional assumption that selecting genotypes of different geographical origin will maximize the diversity available to a breeding project does not hold in sesame.
4. Conclusion
The feasibility of detected genetic variation and relationships using a PCR based strategy was tested in this study. The results indicated variation at the DNA level in the investigated material using RAPD techniques. Based on cluster analysis of RAPD data, the 15 accessions were classified in two major groups and one independent accession, Balinzhima. Sesame accessions from different localities and those differing in seed coat colour were clustered together respectively. The relationships depicted in the dendrogram may be supported by the heterogeneity occurred within individual accession. Overall, the results revealed moderate genetic base for sesame collection studied using seed storage protein and esterase markers. It showed relationship between the sesame samples and geographical origin and also suggests common genetic background between the genotypes. The present study can therefore be employed in the selection of genotypes from various sources to form a wide gene pool with broad genetic base on which future breeding project in Benin could be hinged.
Acknowledgements
F. J-B. Quenum thanks the UNESCO-Biochemical Action Council for providing a short term Fellowship during this work.