1. Introduction
The Phage shock protein (Psp) response maintains bacterial membrane integrity during extracytoplasmic stress. The central component of this system is a 25 kDa protein, which was observed during filamentous phage infection in Escherichia coli but was absent in non-infected cells [1] . This protein was named the Phage shock protein A (PspA) and has been shown to be key to the Psp response [2] . Other inducers of the Psp response (and PspA) include heat shock, hyperosmotic shock, the presence of ethanol [3] , alkaline conditions at stationary phase (pH 9.0) [4] , exposure to hydrophobic solvents [5] , exposure with inhibitors of lipid biosynthesis (i.e. diazaborane and cerulenin) or proton ionophores (i.e. carbonylcyanide m-chlorophenylhydrazone) [4] and membrane stresses related to secretion [6] -[9] .
The Psp system in E. coli contains three divergent transcriptional units. The first is regulated by an upstream s54-promoter and contains five open-reading frames encoding: PspA, PspB, PspC, PspD and PspE proteins (referred to hereafter as the psp operon) [10] . The second transcriptional unit encodes the bacterial enhancer-binding protein PspF, which acts as a transcriptional activator of the psp operon [3] [11] . The third gene is not linked to the psp operon and pspF, but is up-regulated in tandem with the psp operon genes by s54, PspF, the integration host factor and PspA; this gene was subsequently named pspG [12] [13] .
PspA is a dual-function protein whose subcellular location is at the interface between the cytoplasm and the inner membrane [14] . Interacting with PspF, PspA forms an inhibitory dodecameric complex that represses transcription of the psp operon [15] -[17] . PspA also forms higher-order species that have a putative role in maintaining cell membrane integrity [18] and restoring the proton motive force [2] . Large ring complexes containing thirty-six PspA sub-units have been identified by electron microscopy and single particle analysis, with each unit having an outer diameter of 20 nm [19] . In addition to these ringed complexes, PspA also assembles into spherical or prolate spheroidal particles of about 30 - 40 nm in diameter that form clathrin-like scaffolds [20] . It has been suggested that these higher-order species observed in vitro also form in E. coli, interacting with the membrane, and restoring cell membrane integrity [18] .
Analogous ring-shaped structures have also been observed with the Bacillus subtilus PspA homologue LiaH, a protein involved in cell envelope stress [21] . The vesicle-inducing protein of plastids (Vipp1) [22] [23] from Chlamydomonas reinhardtii, also assembles into similar ring-like structures [24] , which have been shown to further assemble into rod-like complexes when purified from Synechocystis sp. PCC 6803 [25] . Vipp1 and PspA are functionally analogous, with Vipp1 able to complement the export defect brought on by a pspA-null mutant strain of E. coli during protein export through the twin-arginine transporter export machinery [8] . However, the reverse was not possible within cyanobacteria that contain both genes [26] . SynechocystisVipp1 and E. coli PspA share 27% sequence identity and a sequence similarity of 51% [27] ; they both contain large regions of putative coiled-coils when analysed with a structural prediction program, with Vipp1 containing an additional α-helix at the C-terminal [26] . Both proteins contain four domains; in E. coli all four domains are required for the formation of the 36-meric species, deletion of domain 4 limits PspA to dimer formation, while the removal of both 1 and 4 leads to hexamer formation [15] . Previous studies have also suggested that the presence of detergent in the protein purification buffers has a significant effect on the type of higher-order species formed by PspA [19] [20] .
PspA was shown to form 36-mer ring-like structures in the presence of 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS, a zwitterionic detergent used to solubilise membrane proteins), in the extraction buffer [19] . Whereas in the absence of CHAPS, larger clathrin-like scaffolds were observed, which were not observed in the presence of CHAPS [20] . It has therefore been suggested that the use of detergent modifies the PspA structure resulting in the observed structural alterations [20] . Regardless of the presence or absence of detergents in the protein purification buffers, the rod-shaped structure observed for Vipp1 (purified in the absence of detergent) have not been previously observed for PspA [25] . Here we report that E. coli PspA also forms the higher-order rod-like species previously observed in Vipp1, regardless of the presence or absence of detergent during protein purification.
2. Materials and Methods
2.1. Strains and Plasmids
E. coli strain DH5α (E. coli Genetic Stock Center) was used for all cloning steps and BL21 (DE3) (Invitrogen, UK) was used for protein expression. The gene encoding PspA (669 bp) was amplified by PCR using the primers BamHI-PspA-F (5’-GCCGCCGGATCCATGTCGCTTTTCGACTCTATTTCGCGC-3’) and PspA-SacI-R (5’- CGGCGGGAGCTCTTACTGCGCCGGCGTGTTCAGTTGC -3’) and cloned into the corresponding sites of pET28a plasmid (Novagen, UK). The resulting pET28-His6-pspA plasmid encodes an N-terminal hexa-histidine tagged PspA protein (His6-PspA). The accuracy of this construct was confirmed by DNA sequencing (Eurofins MWG, Germany).
2.2. Expression and Purification of PspA from Escherichia coli
The expression of His6-PspAwas carried out in E. coli BL21 (DE3) cells, as previously described [15] , with or without 1.1% CHAPS in the buffers. Briefly, pET28-His6-PspA was transformed into chemically-competent BL21 (DE3) cells. These cells were subsequently grown in LB medium to an OD600 of approximately 0.6, and protein expression was induced with 1 mM IPTG and incubated further overnight at 18˚C. After incubation cells were harvested by centrifugation. Cell were resuspended in lysis buffer (100 mM Tris-HCl (pH 7.5), 50 mM NaCl, 75 mM NaSCN) and lysed by sonication. The insoluble and soluble fractions were separated by centrifugation. The insoluble fraction was extracted with extraction buffer (100 mM Tris-HCl (pH 7.5), 600 mM NaCl, 75 mMNaSCN, with and without 1.1% CHAPS) for 2 hours at 4˚C. Fractions were separated by centrifugation and then purified by nickel-affinity chromatography (as per manufacturer’s protocols) using a wash buffer (100 mM Tris-HCl (pH 7.5), 50 mM NaCl, 75 mM NaSCN, 20 mM imidazole) and eluted from the column by the addition of 500 mM imidazole. When the experiment was carried out in the presence of CHAPS, 1.1% was added to the elution solution.
2.3. Size Exclusion Chromatography
Affinity-purified His6-PspA was further purified by gel filtration on a Superdex 200 gel filtration column (GE Healthcare, UK) using a flow rate of 1 ml/min at room temperature in gel filtration buffer (20 mM Tris-HCl (pH 7.5), 200 mM NaCl, 75 mM NaSCN). Fractions (1 ml) were collected between 40 and 52 ml. The column was calibrated with the molecular mass standards of blue dextran (2000 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), albumin (66 kDa), carbonic anhydrase (29 kDa) and cytochrome C (12.4 kDa), all in gel filtration buffer (MWGF200, Sigma-Aldrich).
Proteins were separated by 12% SDS-PAGE and transferred onto nitrocellulose transfer membranes (Protran, Whatman). The blots were blocked for 1 hour in 5% milk in PBS with 0.05% TWEEN at room temperature. They were then incubated for 1 hour with anti-His (27-4710-01, GE Healthcare) at 1:1000 dilution in 5% milk in PBS with 0.05% TWEEN followed by incubation with anti-mouse-HRP (NA931, GE Healthcare) at 1:5000 dilution. Secondary antibody was detected using an enhanced chemiluminescence reagent (Western C, Biorad).
2.4. Electron Microscopy
Protein samples were concentrated using Pierce Concentrator 20 K MWCO (Fisher Scientific, UK). Samples of purified PspAwere negatively stained with 2% (w/v) uranyl acetate on 50% carbonyl formvar grids, before imaging on a Hitachi H7000 Transmission Electron microscope operated at 80 kV with magnifications ranging from ×50,000 to ×200,000.
3. Results
3.1. Purification of the Protein
His6-PspA was purified from the insoluble fraction of E. coli BL21 (DE3) using a previously reported method [15] . To assess the role of detergent on the structure of PspA, purification was carried out in the presence or absence of 1.1% CHAPS. Both approaches yielded pure His6-PspA as observed on SDS-PAGE (Figure 1(A)). To determine the molecular mass of the His6-PspA species, the nickel-affinity purified products were analysed by gel filtration (Figure 1(B) and Figure 1(C)). For the protein sample purified in the presence of CHAPS, the PspA protein eluted at 43.4 ml and between 67.3 - 108.7 ml, while PspA purified in the absence of CHAPS eluted at 44.3 ml and 59.7 - 109.4 ml. The peak eluting from both samples at approximately 44 ml corresponds to a PspA species with a molecular mass greater than 200 kDa; these have been shown to include the putative 36-meric ring structure and may include other higher-order PspA complexes. The samples eluting between 59 - 110 ml correspond to hexameric (calculated at 68.0 ml), dimeric (calculated at 81.9 ml) and monomeric (calculated at 90.3 ml) species. The presence of His6-PspA in the peak corresponding to higher molecular weight species at 44 ml was confirmed by Western blot analysis (Figure 1(D) and Figure 1(E)).
3.2. Visualisation of PspA
Previous studies have indicated that the function of PspA in the Psp response is dependent on its oligomerisation [18] . Given that the PspA homologue Vipp1 assembles into rod-like structures, we hypothesised that the previously observed ring-like 36-meric PspA complexes would also self-assemble into rod-like superstructures. We probed this hypothesis using electron microscopy; purified His6-PspA (eluted in fractions at 43 - 44 ml) was
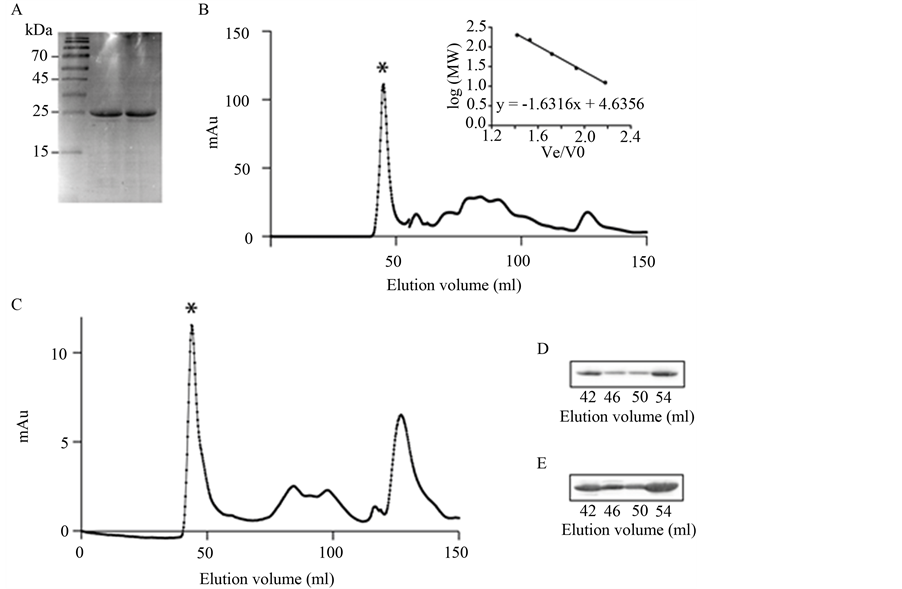
Figure 1. Purification of PspA protein. (A) SDS-PAGE of His6-PspA purified in the absence of CHAPS detergent (lane 2) and in the presence of CHAPS detergent (lane 3). Lane 1 is ladder (Fermentas Page Ruler, PN26616, Fisher Scientific); (B) Elution profile for His6-PspA purified in the presence of CHAPS detergent. Peaks at 43.4 ml (*) and a range between 67.3 - 108.7 ml correspond to higherand lower-order complexes, respectively. Insert shows gel filtration calibration (R2 = 0.9974); (C) Elution profile for His6-PspA purified in the absence of CHAPS detergent. Peaks at 44.3 ml (*) and a range between 59.7-109.4 ml correspond to higherand lower-order complexes, respectively; (D) Western blot of His6-PspA in the oligomeric peak (*) purified in the presence of CHAPS detergent; (E) Western blot of His6-PspA in the oligomeric peak (*) purified in the absence of CHAPS detergent.
herefore absorbed onto formvar grids, negatively stained with 2% uranyl acetate, and imaged. The effect of the presence or absence of detergent (CHAPS) in the purification buffer on the self-assembly of PspA was also assessed by imaging His6-PspA purified in the presence, or absence of CHAPS.
No difference was observed in the structures adopted by His6-PspA purified in the presence of CHAPS compared to that purified in the absence of CHAPS. Both samples showed a mixture of two distinct structures (Figure 2(A) and Figure 2(B)); the first species were the previously observed [19] ring-shaped complexes, which had been postulated to correspond to a 36-meric species of PspA. The outer diameter of these ring-like structures observed (Figure 2(D)) was ~30 nm [19] . A ring of weak contrast with a black, stain-filled region in the middle is clearly observed, suggesting a low protein concentration and thus a hole in the centre with a diameter of approximately 7 - 10 nm, consistent with previously reported data [19] .
The second type of structure observed in both samples was rod-like (Figures 2(A)-(C)), analogous to the rodlike complexes observed for Vipp1 [25] . The diameter of the Vipp1 rod-like complexes averaged between 30 and 40 nm, in agreement with the diameter of the rod-like structures observed here. The average length of the rods was around 500 nm, although significantly longer and shorter species were frequently observed (Figure 2(A) and Figure 2(B)). Along each rod-like complex striations and indentations were visible (Figure 2(C)), even at lower magnification. These were uniform and approximately 30 nm apart (Figure 2(D)) suggesting that they were composed of the super-assembly of the ring-like structures stacked on top of each other; similar striations and indentations are observed with Vipp1 [25] . These rod-like structures were observed to be both curved and straight, and to have open or tapered ends (Figures 2(A)-(C)).
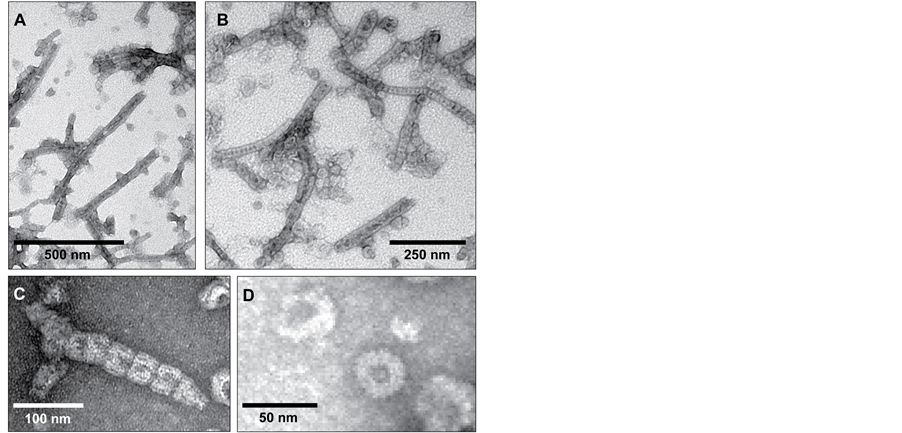
Figure 2. Electron micrographs of negatively-stained PspA complexes. (A and B) A mixture of ring-like and rod-like structures are readily visible; (C) Close-up of a rod-like structure, clearly showing the indentations and striations that indicate stacking of ring-like structures. This is also an example of the tapered end observed occasionally; (D) A ring-like PspA structure. Magnifications range from ×50,000 to ×200,000.
It should be noted that none of these species were aggregated protein; within both samples protein aggregation was observed (not shown), however, this was minimal (approximately 2% - 5%) and gave a disordered mass that was distinct from the self-assembled species depicted here. The super-assembly of E. coli PspA protein into these ordered, rod-like structures indicates that the previously reported ring-like structures are not the final form of PspA. Our data suggests that these ring-like structures further interact to form rod-like species, with a physiological role (Figure 2(A) and Figure 2(B)).
4. Discussion
We have demonstrated by electron microscopy that E. coli PspA units self-assemble to form rod-like complexes analogous to those previously observed for C. reinhardtii Vipp1 [24] . Both proteins (Vipp1 and PspA) are predicted to be highly α-helical (97%), with the α-helical domains in Vipp1 demonstrated to be critical to the formation of higher-order complexes [28] . We found that the formation of these rod-like complexes was not dependent, or affected by the presence or absence of detergent in the purification buffer (as previously hypothesised) [20] . Several studies had previously reported the formation of ring-like [19] and clathrin-like structures [20] for E. coli PspA, but this is the first report of the formation of rod-like structures for this protein. One possibility for the absence of this species in prior studies could be due to the concentration of the protein used. Studies have shown that when Vipp1 expression is significantly reduced (due to the use of a mutant strain), these rod-like structures are no longer observed [29] . This suggests that protein concentration is a key determinant for the formation of these higher-order PspA structures. Once formed, these higher-order structures likely restore membrane integrity by localising to the cytoplasmic face of the inner membrane; PspA is known to interact with the phospholipids phosphatidylglycerol and phosphatidylserine when in its oligomeric form [18] . It may be postulated that these rod-like structures are the physiologically relevant form of PspA, functioning as scaffolds to maintain membrane integrity during phage shock protein response to membrane stress.
Additional evidence for the biological relevance of the observed PspA rod-shaped species comes from a study that utilised GFP-fused PspA to monitor the subcellular location of this protein before and after induction. PspA was found to be evenly distributed within the cytoplasm prior to induction, with highly dense clusters observed at the membrane (primarily around the poles) after PspA induction [14] [30] . The large variation in the fluorescence intensity of the GFP-PspA clusters is characteristic of the formation of several higher-order species; 59% of the observed clusters showed a twoto three-fold increase in intensity above background GFP, while the intensity of a further 20% of the clusters had increased by more than 6-fold [14] . In light of our observations, it is possible that clusters showing a large increase in fluorescence intensity correspond to the rod-like species, while the species showing lower fluorescence intensity may be the 36-meric ring species or clathrin-like scaffolds, previously observed [19] [20] . It should be noted that such localisation has also been observed with Vipp1 [25] . It is interesting to note that the analogous rod-shaped complexes formed by Vipp1 are not permanent, but have been found to be disassembled by HSP70B(DnaK)-CDJ2-CGE1 chaperones in vitro; when formed in cell extracts, these structures are also disassembled by the chaperones in an ATP-dependent process [24] . Thus it may be envisaged that the mesh-like scaffolds formed by PspA are dynamic structures that are disassembled by an analogous, currently unknown mechanism.
The self-assembly of ring-shaped protein complexes to rod-like structures has also been shown with the PspA homologue LiaH [21] , and with holins [31] . Oligomerisation of holins forms lethal holes in the cytoplasmic membrane, thus controlling the length of the infection cycle of tailed phages. Both LiaH and the holin protein S105 have been shown to have the potential to assemble into higher-order structures [21] [31] . Higher-ordered PspA species have been shown to be required for the interaction with liposomes in vitro [18] , but none of these species (ring-like, clathrin-like or rod-like structures) have been observed in bacteria, therefore it is possible that they may simply be an artefact arising in vitro. Thus one can only speculate as to the physiological role of the observed rod-like complexes; the biological implications and importance of these structures is unknown and further studies are required to better elucidate their purpose and function.
NOTES
*Corresponding author.