Application of Graph Entropy in CRISPR and Repeats Detection in DNA Sequences ()
1. Introduction
Deciphering the enormously long nucleotide sequences that are being uncovered in the human genome is one of the major challenges in our days. Along with serious ethical issues, we encounter a series of tremendously hard scientific problems. These problems mainly arise from the fact that although sequencing techniques are almost completely automatic controlled the analysis of the sequenced data is not. Hence, the major goal of the Human Genome Project is the extraction of biologically and medically relevant information from almost automatically sequenced DNA and RNA molecules. In principle, biochemical methods are able to do this job, but since they are extremely expensive and time consuming, there is a high demand for alternative approaches to extract the information hidden in genome [1] . In this situation, concepts and techniques from information theory turned out to be welcoming tools to handle the problem of extracting valuable information from biosequences such as DNA, RNA, or amino acid chains. The main goal of our work is the presentation of a concept and method derived from information theory that will apply to problems of analysis of DNA.
The motivation for this study is to analyze DNA sequences to determine interesting sections of genome that has repeating features using information theory tool.
In many organisms, the genomic DNA is highly repetitive accounting for close to 5% of the genome size [2] [3] . Repetitive DNA sequences are a major component of eukaryotic genomes and may account for up to 90% of the genome size [4] . The human genome itself has over two-thirds of the sequence consisting of repetitive elements [5] . The identification of repeats has proven to be of significance, as they provide insight into the functional and evolutionary roles of various organisms [6] [7] [8] [9] [10] .
In our study we also focus on a family of repeats known as Clustered Regularly Inter Spaced Palindromic Repeats (CRISPRs) [11] . CRISPRs have attracted a great deal of interest recently in genome editing [12] . CRISPRs have been found only in the genomes of prokaryotes and are composed of short direct repeats currently known to range in sizes from 21 - 47 base pairs. This family of repeats is unique in that they are interspaced by non-repeating sequences of similar size, called spacers. CRISPRs were found in approximately 40% of bacterial genome investigated [13] .
Several software applications are available for identifying various form of repeats in [14] [15] [16] .
2. Graph Entropy Algorithm
A graph is an object that consists of a non-empty set of vertices and another set of edges. Vertices are often called nodes, and edges are referred as connections. The set of edges may be empty, in which case the graph is just a collection of points.
We say that two vertices i and j of a directed graph are connected if there is an edge from i to j or from j and i. Suppose we are given a directed graph with n vertices. We construct an n × n adjacency matrix A associated to it as follows: if there is an edge from vertex i to vertex j, we put 1 as the entry on row i, column j of the matrix A; if there is no edge, we put 0.
If one can walk from vertex i to vertex j along the edges of the graph then we say that there is a path from i to j. If we walked on k edges, then the path has length k. For matrices, we denote by Ak the matrix obtained by multiplying A with itself k times. The entry on row i, column j of A2 corresponds to the number of paths of length 2 from vertex i to vertex j in the graph.
Let us consider a directed graph and a positive integer k. Then the number of directed walks from vertex i to vertex j of length k is the entry on row i and column j of the matrix Ak, where A is the adjacency matrix.
In this section, we will discuss entropy of such adjacency matrix A. Let 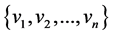 be a set of vertices of a direct graph. Let
be a set of vertices of a direct graph. Let 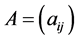 be the
be the 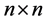 adjacency matrix with at least one positive element.
adjacency matrix with at least one positive element.
Let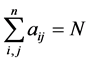 . Let
. Let 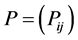 be the matrix such that
be the matrix such that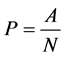 . Hence
. Hence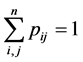 .
. 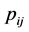 is the probability of having a path from vertex
is the probability of having a path from vertex 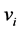 to vertex
to vertex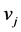 . Adding all elements of each row of P and placing them on the diagonal, we form a diagonal matrix
. Adding all elements of each row of P and placing them on the diagonal, we form a diagonal matrix
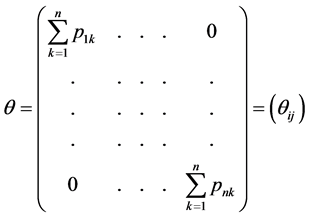
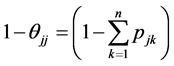 is the probability for a randomly generated path to end at the vertex
is the probability for a randomly generated path to end at the vertex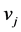 . Let
. Let 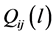 be the probability for generating a path of length l that begins at
be the probability for generating a path of length l that begins at 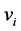 and ends at
and ends at 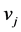 for any integer l. For example, we have
for any integer l. For example, we have 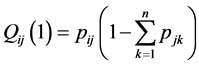 and
and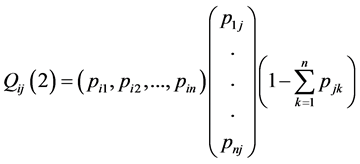 . Let
. Let ![]() be the matrix whose ij element is
be the matrix whose ij element is![]() . Then we have
. Then we have![]() . Finally, we define the asymptotic walk matrix
. Finally, we define the asymptotic walk matrix ![]() as
as![]() , Where
, Where ![]() is the probability for generating a walk of any length from
is the probability for generating a walk of any length from ![]() to
to![]() .
.
Note that ![]()
![]()
We noticed that the sum of all entrees of the matrix![]() , for any integer
, for any integer![]() , is 0. Since sum of all entrees of P is 1, sum of all entrees of
, is 0. Since sum of all entrees of P is 1, sum of all entrees of ![]() is also1. We therefore define the asymptotic entropy
is also1. We therefore define the asymptotic entropy
![]()
where ![]() is defined to be 0 for
is defined to be 0 for![]() . This can also be called the graph entropy of the graph or entropy of the adjacency matrix A. For illustration, Let us consider a short sequence:
. This can also be called the graph entropy of the graph or entropy of the adjacency matrix A. For illustration, Let us consider a short sequence:
ATGCCTGATGCGACGC
Taking 2-letter nodes with one overlap, we can create a graph as following:
![]()
We draw a graph as in the Figure 1.
For our sequence, graph entropy
![]()
3. Results
We have downloaded wide range of genome data, eukaryotes (animals, plants, insects, fungus) and prokaryotes (bacteria, archaea) from Gen Bank: ftp://ftp.ncbi.nlm.nih.gov/genomes/.
We have implemented the Graph Entropy Algorithm in MATLAB platform and converted data to MATLAB format. Then we have computed graph entropy using our Graph Entropy Algorithm by scanning the data with a typical sample size of 512 base pairs (bp) and step size of 10 bp taking 3 nodes with 1 overlap. We have drawn graphs of entropy versus genome length of Acidovorax bacteria in Figure 2, Salmonella-Typhi CT18 bacteria in Figure 3, Caldicellulosiruptor Kristianssonii bacteria in Figure 4 and Human Chromosome-21 in Figure 5. We have studied the intervals visually where entropy was low and found some repetitive pattern in the sequence. Once we have a string of repetitive pattern we used MATLAB “strfind” command to find out exact positions of the repetitive patterns. We have included few examples in this paper, only the ones we thought important.
In Figure 2 we looked at the lowest drop of entropy which is at: x: genome length = 871,100, y: entropy = 4.088. We took an interval (871,000, 871,600) around the lowest drop x = 871,100.
![]()
Figure 2. Acidovorax (bacteria) genome length vs entropy.
![]()
Figure 3. Salmonella-typhi CT18 (bacteria).
![]()
Figure 4. Caldicellulosiruptor kristianssonii (bacteria).
The following is the sequence in the interval taken. The colored string is repeating.
ATAAAAAAACCCGGTGCATGCACCGGGTGGGACCAGCCCCGCGGGCGGGGCGGCTGGCTGCTGTCGTCGCTCAGGGCTTGGTGCCCGTCGGGAAGGGCCATGCGGCCTGCGGGTTCAGCGTGGTCTGTGCTGCGGGTGCAGGCGCAGGGGCAGAGGCCTTGGAGGCCGCCTTTTTCGGGGCAGCCTTCTTCGGTGCAGCGGCCTTGGTCGTGCCGGTGGCCTTCTTCGCCGGTGCAGCTGCCTTCTTGGTGGAGGCTGCGGCCTTCTTTGCCGGTGCAGCTGCCTTCTTGGCGGGGGCTGCGGCCTTCTTCGCCGGTGCAGCTGCCTTCTTGGCGGGGGCTGCGGCCTTCTTTGCCGGTGCAGCTGCCTTCTTGGCAGGAGCTGCGGCCTTCTTTGCCGGTGCAGCTGCCTTCTTGGCGGGGGCTGCGGCCTTCTTTGCCGGTGCAGCTGCCTTCTTGGCAGGAGCTGCGGCCTTCTTTGCCGGTGCAGCTGCCTTCTTGGCAGGAGCTGCGGCCTTCTTTGCCGGTGCAGCTGCCTTCTTGGCGGGGGCTGCAGCCTTCTTCGCCGGAGCGGCCTTCTTCGTCGTGGCGGCGGCCTTCTT
Strfind(g,'GCCGGTGCAGCTGCCTTCTTGG') command gave us the following positions of those repeats in the sequence.
871227 871269 871311 871353 871395 871437 871479 871521
The spacers are almost identical. These are tandem repeats.
Similarly in the Figure 3 we looked at the lowest drop of entropy which is at x = 2926000 y = 4.923.
We looked at the DNA sequence in the interval (2926000:2926650) around x = 2926000. The following is the sequence in the interval taken. The colored string is repeating.
AAAAATGCATCCTTCCCGAACGGCAATAGCTGGCACGACGTACGGCTTGATAATCAACAGCATATAGACAAGGCGCTGCCAGGGCGGATTGAGCGCCGTAGCCGCGATGTAGTGCGGATAATGCTGCCGTTGGTAAAAGAGCTGGCGAAGGCGGAAAAAACGTCCTGATATGCTGGTGAAACGTGTTTATCCCCGCTGGCGCGGGGAACACGGACAGCAACCCGTGTCGGATATCAGACAGATCGGTTTATCCCCGCTGGCGCGGGGAACACACGCGAATCGCCAATCGCCGCCGCGTGAATTGCGGTTTATCCCCGCTGGCGCGGGGAACACCCACGATGTATGCCGACCGTGATTTTTACCGCCGGTTTATCCCCGCTGGCGCGGGGAACACAGATACGCCTTTACGTCGCCCTCTTTGGCGCGCGGTTTATCCCCGCTGGCGCGGGGAACACTAAAACACCGGTTGCGCAACCTCCGCGGGGATCGGTTTATCCCCGCTGGCGCGGGGATCGGTTTATCCCCGCTGGCGCGGGGATCGGTTTATCCCCGCTGGCGCGGGGAACACTCTAAATCTACCCAATTGAATTTAAATACTTTTTTAGCGCACAAAAAACCCACCAACTTTTCCTAATTTTTAAAGATCTCTAA
We used strfind(g,'CGGTTTATCCCCGCTGGCGCGGGGAACAC') in MatLab and found more repeats outside the interval.
2926243 2926304 2926365 2926426 2926539 2943184 (does not belong to this region). length('CGGTTTATCCCCGCTGGCGCGGGGAACAC')=29 strfind(g,'GTGTTTATCCCCGCTGGCGCGGGGAACAC'): 2926182 strfind(g,'CGGTTTATCCCCGCTGGCGCGGGGATCGG') 2926487 2926513.
Starts: 2926182 Ends: 2926567.
In the interval (2926182, 2926513) we find three strings differing by 2 to 4 letters.
These repeats are called CRISPR. This is only CRISPR so far known for this strain of the bacteria.
Again, we studied the pattern of the DNA sequence of Caldicellulosiruptor Kristianssonii (Bacteria) in intervals around the points of low entropy and found repetitive patterns. In Figure 4, we considered the drop at x = 2,672,000, y = 4.46. Following is the sequence in the interval (2671900, 2672600) around this drop. The repeats are shown in red color.
TATTGCAATTATTGTCCTATGCACAGAGTTTGTAGCCTTCCCGTTGGGGATTGAAACATAGATTTCATTTCGCAGCCAATAGAGCGGTTTATAGTTTGTAGCCTTCCCGTTGGGGATTGAAACCTCAATTTCTGTTTCTCTTTTCTCAATTATTCTTGAGTTTGTAGCCTTCCCGTTGGGGATTGAAACTATAATAGCCCATTCATCAAAAACTTTTTCATCGAAGTTTGTAGCCTTCCCGTTGGGGATTGAAACTATAATAGCCCATTCATCAAAAACTTTTTCATCGAAGTTTGTAGCCTTCCCGTTGGGGATTGAAACCACAAAATTATAGTTTGGCGCAATGTAAACACGAACAGTTTGTAGCCTTCCCGTTGGGGATTGAAACTCTATGTCTTCTTCAAGATACATATCGAGCAGCTTATTGTTTGTAGCCTTCCCGTTGGGGATTGAAACATACTTTTTTTCTCACGGTCTGTATGGCCTGTTCAGT
We notice repeats and use Matlab to find the exact locations of that string.
strfind(g,'GTTTGTAGCCTTCCCGTTGGGGATTGAAAC')
Columns 1 through 61
2666352 2666419 2666484 2666551 2666616 2666682 2666748 2666813 2666879 2666947 2667014 2667081 2667147 2667213 2667278 2667344 2667410 2667476 2667544 2667611 2667676 2667741 2667805 2667872 2667939 2670817 2670882 2670949 2671016 2671081 2671147 2671214 2671279 2671345 2671411 2671476 2671544 2671610 2671675 2671740 2671805 2671871 2671936 2672001 2672070 2672135 2672201 2672267 2672333 2672399 2672466 2672534 2672599 2672666 2672733 2672798 2672864 2672930 2672996 2673523 2673590
Length ('GTTTGTAGCCTTCCCGTTGGGGATTGAAAC')=30
61 repeats of length of 30, unique spacers
Starts: 2666352 Ends: 2673620 Period: 65/66/67 Total Length = 7268
These repeats are CRISPR.
In Figure 5, we considered the drop at x = 44010000, y = 4.13 and the interval (44009900, 44010500). Following is the sequence of Human Chromosome-21in that interval. We also found repeats.
CCGTTTATATCCACGCAGGCGTTTCCCCTTACCTGCACCGAGCCTCCATTCCCGTTTATATCCACGCAGGCGTTTCCCCTTACCTGCACCGAGCCTCCCGCCCCGTTTACATCCACGCAGGCGTTTCCCCTTACCTGCACCGAGCCTCCATTCCCGTTTATATCCACGCAGGCGTTTCCCCTTACCTGCACCGAGCCTCCCGCCCCGTTTACATCCACGCAGGCGTTTCCCCTTACCTGCACCGAGCCTCCATTCCCGTTTATATCCACGCAGGCGTTTCCCCTTACCTGCACCGAGCCTCCCGCCCCGTTTACATCCACGCAGGCGTTTCCCCTTACCTGCACCGAGCCTCCATTCCCGTTTATATCCACGCAGGCGTTTCCCCTTACCTGCACCGAGCCTCCATTCCCGTTTATATCCACGCAGGCGTTTCCCCTTACCTGCACCGAGCCTCCATTCCCGTTTATATCCACGCAGGCGTTTCCCCTTACCTGCACCGAGCCTCCCGCCCCGTTTATATCCACGCAGGCGTTTCCCCTTACCTGCACCGGGCCTGCCGCCCCGTTTACATCCACGCATGCGTTTCCCCTTACCTGCACTG
strfind(g,'TTTCCCCTTACCTGCACCGAGCCTCCATTCCCGTTTATATCCACGCAGGCG')
Columns 1 through 18
44007626 44008952 44009105 44009156 44009258 44009360 44009462 44009513 44009615 44009717 44009819 44009870 44009921 44010023 44010125 44010227 44010278 44010329
The spacers are almost identical with this string except 4 letters (in purple). We also find the spacer string.
Strfind(g,“TTTCCCCTTACCTGCACCGAGCCTCCCGCCCCGTTTACATCCAC GCAGGCG”).
Columns 1 through 23
44007575 44007677 44007728 44007779 44007830 44007881 44007983
44008034 44008289 44008493 44008646 44008697 44008799 44008850
44008901 44009207 44009309 44009564 44009666 44009768 44009972
44010074 44010176
This is a repeat of a string without any gap in the region (44007575, 44010329).
Discussion
The importance of identifying repetitive sequences is clear; however, the considerable size of many genomes makes fast and efficient repeat detection very challenging. In this paper, we have presented a new algorithm for finding repeats in DNA sequences. The algorithm is based on our new measure of entropy for any non-trivial graph. In [15] , an algorithm were presented for finding tandem repeats in DNA sequences based on the detection of k-tuple matches. It uses a probabilistic model of tandem repeats and a collection of statistical criteria based on that model. Whereas in [14] and [16] a new tool was introduced for the automatic detection of CRISPR elements in genome. The main advantage of our tool is it will detect both tandem repeats and CRISPR or any other repeats. The main disadvantage of our tool is lack of complete automation and hence it is less efficient compared to the other tools. Our detection technique convert sequences to an alternative representation (namely, graph as it is given in [17] ) in an attempt to make analysis more efficient. Future research plans are to modify the presented algorithm so that it is also able to identify repeats efficiently. Our code will be available to the reader upon request through email to one of the authors.
4. Conclusions
We have studied the following species:
Eukaryotes: Homo sapiens chromosome 19 & 21, Anopheles gambiae, Caenorhabditis elegans, Plasmodium falciparum Saccharomyces cerevisiae.
Prokaryotes: Acidovorax, Ammonifex, Caldicellulosiruptor kristjanssonii, E.Coli, Salmonella Typhi, Listeria Monocyto genes, Bacillus clausii KSM, Chlamydia muridarum Nigg, Cyanobacterium aponinum, Gluconacetobacter diazotrophicus, Haemophilus influenzae R2866, Mycobacterium tuberculosis, Mycoplasma genitalium, Neisseria meningitidis, Streptococcus pneumoniae, Thermosipho africanus, Truepera radiovictrix (Bacteria), A. fulgidus (Archaea).
Viruses: HIV, Hepatitis B. After analyzing the DNA sequence at the points of low entropy for all these species, we conclude that low entropy in a genome graph corresponds to high repeatability in the sequence. These repeats can be classified as CRISPR or Tandem Repeats or something else.
Acknowledgements
This paper was written while two authors were Summer Faculty Fellow in SPAWARS YSCEN Atlantic, Charleston, SC funded by Office of Naval Research. Authors are thankful to their mentor for his assistance in the work.