Differential Regulation of Proteins and a Possible Role for Manganese Superoxide Dismutase in Bioluminescence of Panellus stipticus Revealed by Suppression Subtractive Hybridization ()
Received 27 April 2016; accepted 30 July 2016; published 2 August 2016

1. Introduction
Approximately 71 species of luminous fungi have been recognized distributed among four evolutionarily distinct euagaric lineages [1] . All known bioluminescent fungi are associated with decaying wood or plant litter and produce extracellular enzymes able to break down lignin to assimilable compounds [2] - [4] . Although fungal luminescence has been noted since ancient times, there were few studies on the genetics of mushroom luminescence until relatively recently. Ruth Macrae was a pioneer on the genetics of luminescence in Panellus stipticus, establishing the hereditary nature of luminescence and also showing that luminescence is dominant [5] [6] . The observations by Macrae were confirmed later by Lingle et al. [7] , who determined that three separate naturally- occurring mutations lead to loss of bioluminescence and also found the existence of non-luminous monospore cultures able to produce luminous dikaryotic mycelium as a result of genetic complementation. Earlier work with bioluminescent Mycena species by Bothe [8] , as cited by Harvey [9] similarly found three “series” of hereditary factors, including some of which were inhibitory suggesting that regulatory genes may be involved as well.
The precise molecular nature of fungal bioluminescence remains unresolved today, with the nature of the light emitter, the enzymes involved and the biochemical mechanism not fully resolved. Two alternative concepts for the cellular organization and molecular mechanism of fungal luminescence are currently being investigated. According to the first concept luminescence occurs in the absence of a specialized enzyme by oxidation of the energized state of the luciferin involving an active oxygen species such as the superoxide anion [10] - [14] . Numerous luminescent mushroom species contain chemiluminescent substances capable of emitting light in a Fenton- like reaction, suggesting the involvement of superoxide anion or other active oxygen species in mushroom bioluminescence [11] [12] . Shimomura [15] proposed that fungal bioluminescence involved the one-electron oxidation of the luciferin molecule to form a luciferin radical that is immediately oxidized into peroxides with subsequent decomposition of luciferin peroxide providing the energy for light emission. Noting that bovine Cu/ZnSOD can block luminescence, Shimomura [11] proposed that SOD and the light emitter are compartmentalised inside the fungal cell, isolating SOD activity from the luminescence reaction. This could explain the observation that the disruption of mushroom tissues results in the elimination of bioluminescence as first reported by Buller [16] .
The second concept is based on evidence for the enzymatic nature of bioluminescence, whereby bioluminescence is mediated by a luciferase enzyme acting on the luciferin light-emitter [17] - [22] . Airth and McElroy [17] were first to observe the emission of light in-vitro using hot and cold extracts from bioluminescent fungi. They further separated the cold extract into soluble and insoluble fractions each containing an essential enzyme for light emission suggesting a two-step enzymatic pathway for fungal luminescence. Supportive observations were reported later by Kamzolkina et al. [19] [20] and Kuwabara and Wassink [23] . Kinetic data reported by Oliveira and Stevani [21] further reinforced this conclusion demonstrating that bioluminescence depends on an NAD(P)H-dependent reductase and a luciferase, the latter presumably oxidizing the reduced luciferin intermediate to produce light. Evidence that all four independent lineages of luminescent fungi share comparable luciferins and luciferases was obtained by Oliveira et al. [24] . Recently, Purtov et al. [25] provided evidence that the naturally occurring fungal metabolite hispidine could be the precursor to luciferin in-common to four different lineages of luminescent fungi. Circadian control of luminescence was demonstrated in Neonothopanus garderi by Oliveira et al. [26] with bioluminescence at highest levels at night. The latter authors concluded that mushroom bioluminescence was an evolutionary adaptation to attract insects for efficient spore dispersal.
Comprehensive reviews on the current state of knowledge of fungal bioluminescence have been published recently that reflect the current impasse involving alternate proposals for the mechanism of fungal bioluminescence [1] [21] [27] - [29] . In an attempt to resolve this issue we employed suppressive subtractive hybridization (SSH) in the current study to detect genes involved in mushroom bioluminescence, comparing a luminous North American isolate with a non-luminous European culture of P. stipticus as well as luminous and non-luminous monospore cultures of the North American strain. SSH combines normalization of DNA fragments with exclusion of sequences common to the compared populations to obtain differentially expressed cDNA.
2. Materials and Methods
2.1. Cultures
Presumed dikaryon strains studied were G20.4 (DAOM 242555) a luminous culture isolated on 16 August 2010 from clustered basidiomes on a decaying stump of Betula papyrifera near Almonte, Ontario, Canada, and DAOM F2118 a non-luminous dikaryon strain isolated in 1931 from a stump of Quercus pedunculata in Darmstadt, Germany (leg. Kallenback and Macrae). Dikaryon cultures were isolated by plating portions of the pileus interior onto Difco™ potato dextrose agar (PDA-BD Diagnostics, Sparks, MD, USA). Single-spore isolates of P. stipticus were obtained by careful extraction under microscopic observation of individual germinating basidiospores released from freshly collected pilei suspended over PDA. The following four monokaryon single-spore cultures were studied, all isolated from the same source as G20.4: ssp5 (DAOM 242556) and ssp6 (DAOM 242557) luminous single-spore strains, and ssp7 (DAOM 242558) and ssp9 (DAOM 242559) non-lu- minous strains. All cultures cited are deposited in the culture collection of Agriculture and Agri-Food Canada, ECORC, Ottawa, Canada (DAOM).
2.2. Luminescence
Luminescence (relative units) of 2-wk old colonies growing on PDA in 3.5cm Petri dishes was determined with a GloMax®20/20 luminometer (Promega, Madison, WI, USA). Baseline intensity of the luminometer was 0 - 200 RU. Differences in luminescence between the luminescent and non-luminescent cultures were 5 - 6 orders of magnitude (Table 1).
2.3. cDNA Synthesis and Suppression Subtractive Hybridization (SSH-MOS)
Total RNA was isolated from mycelium harvested from 2-wk old cultures using the Total RNA Isolation NucleoSpin® RNA Plant Kit (Macherey-Nagel, Düren, Germany) using Lysis buffer RAP and following the manufacturer’s instructions. Total RNA was measured with a NanoDrop® ND-1000 spectrophotometer (Thermo- Scientific, Wilmington, DE, USA). For samples with low total RNA (<2 μg), cDNA was synthesized from total RNA using the SMARTer™ Pico PCR cDNA Synthesis Kit (Clontech, Mountain View, CA, USA). For higher concentrations of total RNA, mRNA was isolated using the GenElute™ mRNA Miniprep Kit (Sigma-Aldrich, Saint Louis, USA), and mRNA isolation repeated once to enhance mRNA purification. Bidirectional suppression subtractive hybridization (SSH) was carried out using Clontech PCR-Select™ cDNA Subtraction Kit (Clontech) following the manufacturer’s directions, starting with first strand cDNA synthesis for isolated mRNA, and with adaptor ligation for synthesized cDNA. SSH was performed separately on 1) North American (DAOM 242555) and European dikaryon isolates (DAOM F2118)―luminous and non-luminous strains respectively; 2) North American monokaryon isolates ssp5 (luminous) and ssp7 (non-luminous); and 3) ssp6 (luminous) and ssp9 (non-luminous) isolates.
![]()
Table 1. Luminescence of two-week old cultures of Panellus stipticus.
1Single spore isolates derived from DAOM 242555; 2European isolate in herb. DAOM; 3North American isolate in herb. DAOM.
2.4. Sequencing and Gene Identification
Subtracted cDNA was cloned with the TOPO TA Cloning® kit (Invitrogen, Carlsbad, CA, USA) using One Shot® TOP10 electrocompetent cells, and transformation performed by electroporation with a Bio-Rad Gene Pulser Xcell™ electroporator in 0.1 cm cuvettes at 1.8kV. Amplification of cloned cDNA was performed with the PCR-Select™ primers (M13F, M13R) using a Labnet Multigene thermocycler (Labnet Intl., Edison, NJ). PCR reactions were prepared in 10μL volume containing the following mix: 1 μL 10X Titanium Taq Buffer (Clontech, Mountain View, CA), 0.5 μL 2 mM dNTP, 0.32 μL each of 5 μM upper and lower primers, 7.76 μL sterile distilled water and 0.1 μL Titanium Taq Polymerase (Clontech). Sequencing reactions were prepared using the ABI Prism® BigDye™ Terminator reaction kit (v3.1, Applied Biosystems Inc., Foster City, CA) in 10 μL volume and 1/8 dilution using 5X sequencing buffer. The cycle sequencing reaction contained the following mix: 1.75 μL 5X Sequencing Buffer, 0.5 μL BigDye V3.1 Mix, 0.5 μL of 3.2 μM primer (T3 or T7), 6.25 μL sterile distilled water, 1.0 μL (10 - 40 ng) PCR template, and employed the following amplification protocol: 25 cycles each of 30 sec denaturation at 96˚C, 15 sec annealing at 50˚C, and 4 min extension at 60˚C. Sequences were obtained using an ABI Prism 3100 Genetic Analyzer (Applied Biosystems). Sequence segments of vector origin were identified and removed using the program VecScreen (http://www.ncbi.nlm.nih.gov/tools/vecscreen/). The resulting clean gene fragments were identified by blastx analysis (http://blast.ncbi.nlm.nih.gov/) on the GenBank non-redundant protein-sequence database using the conceptually translated nucleotide sequence. Acceptable identifications contained conserved domains and/or had highly significant sequence similarity (E value < 1e−10) to known genes of other basidiomycetes. Sequence fragments representing genes with possible relevance for luminescence were deposited in GenBank and accession numbers cited in Tables 2-4.
2.5. Chemical Determination of the Superoxide Dismutase Metal Cofactor
The following SOD inhibitors were used to determine the metal cofactor of SOD: 5 mM sodium azide, 5 mM hydrogen peroxide and 3 mM potassium ferricyanide according to standard procedures for cofactor determination of Roberts and Hirst [30] , and treatment with 2% sodium dodecyl sulfate modified from Bannister et al. [31] all in aqueous solutions. Since mushroom luminescence evidently occurs at the cell surface [32] , luminescence inhibitors/effectors were tested by application of the above reagents directly to the surface of luminescent mycelium as follows. Mycelium from the luminescent dikaryon strain (DAOM 242555) for chemical inhibition experiments was grown for two weeks in 2.4% potato dextrose broth, pH 5.1 (BioShop Canada Inc., Burlington, ON, Canada). A sample of luminescent mycelium 0.5 - 1.5 cm2 was extracted from the broth culture and placed in a 3.5 cm Petri dish and an initial luminescence measurement taken for 1 - 2 min duration as a baseline measurement using a GloMax®20/20 luminometer. Afterwards, from 50 - 100 μL of inhibitor solution was dropped onto the mycelium to just cover the entire sample surface and luminescence measurements renewed until stabilization. Water was used as control and inhibition experiments were replicated once to confirm results.
![]()
Table 2. Up-regulated proteins1 in North American (DAOM 242555) vs European (DAOM F2118) isolates.
1GenBank accession numbers in brackets; 2sequence extended to N-terminus (KJ842128).
![]()
Table 3. Up-regulated proteins1 in luminous (ssp5) vs nonluminous (ssp7) monokaryons.
1GenBank accession numbers in brackets.
![]()
Table 4. Up-regulated proteins1 in luminous (ssp6) vs nonluminous (ssp9) monokaryons.
1GenBank accession numbers in brackets.
2.6. Sequencing and Subcellular Localization of the SOD Gene Product
The SOD sequence fragment obtained by SSH was extended using primers designed from within the 3’ UTR region of the fragment obtained by SSH and from within the N-terminus region of an alignment of Genbank MnSOD sequences. PCR was performed as above using primers SOD-F (5’-CTC CCY TAC GCC TAY GAT G-3’) and SOD-R1 (5’-AGC TTG GAA CCC TCG ACA A-3’) with the following amplification protocol: 3 min at 95˚C; 35 cycles of (60 s at 94˚C, 75 s at 65˚C, 105 s at 72˚C); 10 min at 72˚C. Sequencing was performed as above using primers SOD-F and SOD-R2 (CC TCG ACA AAG CGT GTC TG) and the sequence deposited in Genbank (KJ842128). Subcellular targeting of the MnSOD was investigated using Target P1.1 (http://www.cbs.dtu.dk/services/TargetP) and SecretomeP 2.0 (http://www.cbs.dtu.dk/services/SecretomeP).
3. Results
3.1. Suppression Subtractive Hybridization (SSH-MOS)
Three bidirectional SSH were performed providing differing although not necessarily contradictory results. The first SSH compared luminescent and non-luminescent dikaryon strains from North America and Europe respectively (Table 2). Increased protein transcript levels were not found in the European strain, possibly a result of the loss of gene function on account of the advanced age of the culture which was originally isolated in 1931. Manganese superoxide dismutase (MnSOD) was up-regulated in the luminous isolate and has important antioxidant properties in eliminating the highly reactive superoxide ion by dismutation. Although up-regulation of MnSOD only in the North American strain could be the result of loss of function in the older European strain, the possibility that the antioxidant/redox recycling properties of MnSOD could impact luminescence was further explored in the current study. Several up-regulated proteins in the North American isolate have regulatory functions. Proteins of the Cop9 signalosome complex regulate the activities of ubiquitin ligases by deneddylation and are specific for removal of NEDD8 post-translational modifications (PTM) of proteins that would otherwise be destined for degradation in the proteosome. Sentrin/sumo-specific proteases are responsible for the deubiqui- tination of targeted proteins by removal of small ubiquitin-like modifiers (SUMOs). RNB domain-containing proteins also may have a role in regulation and gene expression by post-transcriptional control of RNA. An AB- hydrolase from the luminescent isolate could have diverse catabolic activities and the function of the UPF0041 family protein is not known.
The second SSH compared the expressed cDNA in two monokaryon isolates (Table 3). As in SSH1 we observed several up-regulated enzymes that could be involved in PTM of proteins impacting gene function or acting as molecular switches. Oligosaccharyltransferase, up-regulated in the luminous strain, has a role in protein folding thereby modifying the tertiary structure and functional conformation of proteins. Up-regulated proteins in the nonluminous strain with regulatory functions included cullin NEDD8 ubiquitine ligase, which facilitates ubiquitin ligation of proteins targeted for degradation thereby having the opposite activity to Cop9 signalosome up-regulated in the luminous strain in SSH1. Rho GDP dissociation inhibitor acts as a molecular switch regulating rho-protein activity which has multiple cellular functions. Pepidylprolyl isomerase is involved in protein folding and the two phosphatases potentially could participate in the reversible PTM of proteins. Dienelactone hydrolase (DLH), also uploaded in the nonluminous isolate, is part of the β-ketoadipate biodegradative pathway for toxic aromatic compounds which could include quinone derivatives and related products of lignin metabolism. Syntaxin participates in exocytosis while cyclohydrolase and the lipase or esterase up-regulated in the non-luminous strain have unknown activities. Similarly the specific activity of the PRO41-like protein in the luminous train is not known.
The third SSH compared the other two monokaryon strains (Table 4), producing a single identifiable up- regulated protein from the luminous strain and none from the nonluminous strain. E3 ubiquitin protein ligase is the recognition module for the ubiquitination system responsible for signalling specific proteins for PTM or degradation.
It is perhaps not surprising that different results were observed from the three different SSH trials. We might expect only few detectable differences in metabolism of the single-spore monokaryon cultures, which were isolated from clustered fruiting bodies occurring on the same birch stump. However, one or a few genetic differences occurring between monokaryon pairs could lead to a cascade of metabolic differences, compounded by the loss of the mediating effect of paired dikaryon nuclei, and result in the loss of luminescence. Notably, in all three experiments we found differences in transcript levels of enzymes involved in PTM of proteins that would affect metabolism. In each experiment there were differences in the regulation of proteins involved in ubiquitination which can signal degradation, alter location, affect activity and promote or prevent protein interactions. The cop9 signalosome protein up-regulated in the luminous dikaryon isolate has deneddylation activity which is in opposition to the activity of nedd8 ubiquitin protein ligase up-regulated in the nonluminous ssp7 strain. The observation that MnSOD was up-regulated in the North American luminous strain was notable considering the well documented antioxidant activity of SODs in catalyzing the dismutation of superoxide (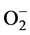 ). Rapp et al. [33] observed that Cu/ZnSOD was capable of forming stable complexes with catechols and other aromatic compounds. Although this ability has not been investigated in MnSOD, the potential for a MnSOD-semi- quinonato complex to react with molecular oxygen or other reactive oxygen species to produce electronically energized products prompted us to consider a possible role for MnSOD in mushroom luminescence.
). Rapp et al. [33] observed that Cu/ZnSOD was capable of forming stable complexes with catechols and other aromatic compounds. Although this ability has not been investigated in MnSOD, the potential for a MnSOD-semi- quinonato complex to react with molecular oxygen or other reactive oxygen species to produce electronically energized products prompted us to consider a possible role for MnSOD in mushroom luminescence.
3.2. Sequencing and Characterization of SOD Up-Regulated in luminescent Panellus
The active cation of the up-regulated SOD in North American luminous Panellus was determined by examining the effect of inhibitors differentially affecting SODs (Figure 1). The different types of SOD are sensitive to different inhibitors―Cu/ZnSOD is inhibited by cyanide and peroxide, FeSOD is inhibited by azide and peroxide, and MnSOD is inhibited by azide and 2% SDS but cyanide and peroxide insensitive [30] [31] . Our experiments showed that luminescence was slightly stimulated by 3 mM cyanide (Figure 1(a)) and was not inhibited by 0.015% H2O2 with luminescence reaching a stable level near the level prior to application after initial stimulation attributable to hydration of the sample (Figure 1(b)). Luminescence was dramatically inhibited by azide (Figure 1(c)) and also inhibited by 2% SDS (Figure 1(d)). These results confirmed that the up-regulated SOD is MnSOD, with inhibition of MnSOD affecting an immediate, significant decrease in luminescence indicative of a possible role for MnSOD in mushroom luminescence. The SOD gene fragment obtained by SSH (GenBank
![]()
Figure 1. Chemical inhibition of SOD in Panellus stipticus. Luminescence is measured in relative units; arrows indicate the time of application of a potential inhibitor of SOD activity: (a) apparent slight stimulation of luminescence on application of 3mM potassium ferricyanide; (b) no inhibition with application of 0.015% H2O2; (c) immediate and complete inhibition of luminescence with application of 5 mM sodium azide; (d) inhibition by 2% sodium dodecyl sulfate (SDS).
JZ516005) contained the C-terminal conserved domain of the Sod_Fe_C family (pfam02777) together with 87 bases of the 3’ UTR. This gene fragment most closely resembled MnSOD in Agaricus bisporus (embl/CAB85688) having 85% similarity with a 2-aa residue modification and 3 residue deletion in the C-terminal conserved domain. The extended cDNA sequence obtained from primers designed for MnSOD comprised 189 aa residues. Protein ligands characteristic of MnSOD were located at positions 26 (histidine), 71 (histidine), 159 (aspartic acid) and 163 (histidine) of the translated protein. The upregulated MnSOD protein sequence from P. stipticus most closely matched a sequence for Laccaria bicolor (XP_001887049) with 85% similarity. Protein sequence similarities were only 53% and 51% with human and Saccharomyces cerevisiae MnSOD respectively, which could indicate functional differences for MnSODs from different organisms. Using the MnSOD primers to sequence genomic DNA resulted in a sequence of 840 bp with three introns at locations 169-217 (49 bp), 407-467 (61 bp) and 606-691 (76 bp) of the alignment. Signal peptides for protein sorting were not detected in the MnSOD sequence; however, non-classical secretion of the protein was predicted in analysis by SecretomeP (SecP score = 0.716).
4. Discussion
4.1. Post-Translational Modification (PTM) of Enzymes Is a Determinant for Luminescence in Panellus stipticus
Proteins involved in ubiquitination were differentially expressed between luminescent and non-luminescent forms in every SSH subtraction, impacting the activities or cellular localization of proteins involved in luminescence. The finding of an extracellular isoform of MnSOD in the luminescent North American form of Panellus stipticus indicates that ubiquitine targeting of this MnSOD to the cell surface in luminescent mushrooms could be a determining factor for fungal luminescence. The association of luminescence with non-mitochondrial membranes was observed by Kamzolkina et al. [19] [20] . Puzyr et al. [32] , using confocal microscopy, observed that luminescence in P. stipticus occurred at the cell surface. Taken together these observations indicate that the luciferase in Panellus is located near the cell surface and associated with the cytoplasmic membrane or cell wall. In eukaryotes MnSOD typically is synthesized in the cytosol and imported post-translationally into the mitochondrial matrix [31] . However, the MnSOD gene in basidiomycetes exists in multiple copies (http://genome.jgi.doe.gov/pages/search-for-genes.jsf?organism=agaricomycotina) potentially expressing functionally different forms [33] - [36] . MnSODs in fungi frequently lack the N-terminus transit-peptide signal targeting to mitochondria and a fungal species may express different MnSODs that are separately located in the cytosol and mitochondria [37] - [40] , with the possibility also for post-translational modification targeting peroxisomes [41] [42] , other cell components or the secretory pathway. Post-translational modifications may impart multiple functions to the same proenzyme in response to differing environmental or physiological conditions (e.g. [43] ). Both the N-terminus and the C-terminus of an SOD proenzyme can contain targeting or retention signals involved in protein sorting. Gould et al. [44] recognized a peroxisomal targeting signal (PTS1) located at the extreme C-terminus comprising the conserved tripeptide sequence SKL. Subsequent studies revealed that at least the last 12 C-terminal residues are involved in modulating peroxisomal targeting efficiency [45] [46] . The PTS1 predictor program developed to recognize PTS1 signal-containing proteins [47] (http://mendel.imp.ac.at/pts1/) did not predict peroxisomal targeting for the MnSOD from Panellus, and signal peptides for protein sorting were not detected in the MnSOD sequence. However, non-classical secretion of the protein was predicted in analysis by SecretomeP (SecP score = 0.716). The possibilities exist, therefore, that the C-terminus of the extracellular MnSOD from Panellus could be modified by addition of a lipid anchor such as glycosylphosphatidylinositol (GPI) allowing insertion into the cytoplasmic membrane [48] [49] , or the enzyme could be targeted for the secretory pathway and retained at the cell surface by ionic interaction with the C-terminal region (e.g. [50] ).
4.2. Could There Be a Role for Extracellular MnSOD in Mushroom Luminescence?
SODs reduce oxidative stress by catalyzing the dismutation of superoxide ion producing H2O2 as represented by the following half-reactions:
Mn3+-SOD + O2∙− → Mn2+-SOD + O2 (1)
Mn2+-SOD + O2∙− + 2H+ → Mn3+-SOD + H2O2 (2)
In human MnSOD removal of O2∙− involves product inhibition and reduction of O2∙− may occur by “prompt protonation” (2) or involve an Mn3-peroxo complex in an “inhibited pathway” (3), (4) which slows catalysis:
Mn2+-SOD + O2∙− → Mn3+(OO2−)-SOD (3)
Mn3+(OO2−)-SOD + 2H+ → Mn3+-SOD + H2O2 (4)
Few organisms have been investigated for product inhibition of MnSOD activity. In the only fungi studied (Saccharomyces cerevisiae, Candida albicans) [51] [52] , superoxide dismutation occurred almost exclusively by the fast protonation pathway. There are significant differences in the ligand backbone between these yeasts and human MnSOD. The aromatic, hydrophobically-neutral, electrically-charged histidine adjacent to the first histidine ligand in human MnSOD is replaced by the aliphatic, branched, highly hydrophobic amino acid isoleucine in the yeasts. In addition, isoleucine adjacent to the aspartic acid ligand in human MnSOD is replaced by the less hydrophobic, unbranched amino acid alanine. The amino acid ligands in the MnSOD of P. stipiticus, as well as those comprising the ligand backbone and the second coordination sphere, were identical or entirely compatible to those in human MnSOD, differing only in the translocation of functionally nearly equivalent valine and isoleucine residues adjacent to the aspartic acid ligand. These observations suggest that SOD dismutation in Panellus may involve the inhibited pathway as seen for human MnSOD.
MnSOD isoforms preferentially bound to cell walls and metabolically distinct from other isoforms may occur widely in eukaryotes [53] [54] where they may act in concert with other cell-wall bound enzymes such as peroxidases [50] [54] . Hydrogen peroxide produced by catalytic dismutation of superoxide ion could be the electron acceptor for the formation of reactive hydroxyl ions by cell wall-bound peroxidases functioning jointly with MnSOD. For example, Kukavica et al. [54] concluded that phenolics associated with the cell wall in pea roots reduced oxygen to superoxide ion in an auto-catalytic manner. The superoxide ion was converted to hydrogen peroxide by a cell-wall-bound isoform of MnSOD which was coupled to a peroxidase generating hydroxyl ion from the hydrogen peroxide produced. In one scenario for fungal luminescence involving MnSOD, extracellular MnSOD catalyzing the dismutation of superoxide and producing H2O2 by the fast protonation pathway could act in concert with a cell wall-bound peroxidase to generate reactive hydroxyl radicals. Hydroxyl radicals could be involved in luminescence by one-electron oxidation of fungal luciferin to produce an excited state resulting in light emission on relaxation. In the following section, however, we propose a hypothetical mechanism whereby the extracellular MnSOD in P. stipticus could participate as the effective luciferase in luminescence.
4.3. A Hypothetical Role for MnSOD as the Fungal Luciferase
In previous studies (unpublished) we observed the darkening of growth media in luminescent cultures of mushroom species (Armillaria borealis, A. mellea, P. stipticus) grown on solid and liquid media, in time producing dark exudates which could be due to the accumulation and polymerization of enzymatically oxidized phenols or quinhydrones. Similar dark metabolites were not observed in non-luminous cultures of the same species grown under the same conditions. This observation suggests a role for the hydroquinone/semiquinone/quinone oxido- reductase triad in luminescence. Rapp et al. [33] observed that ο-hydroquinone as well as a group of substituted catechols can be oxidized in the presence of oxygen to the semiquinone form by reaction with Cu/ ZnSOD while simultaneously reducing the enzyme’s metal cofactor. The resulting semiquinone forms a complex with the enzyme by ligation to the cofactor, stabilizing the semiquinone radical and greatly reducing the rate of radical termination by disproportionation [55] . Non-enzymatic electron transfer from a reactive oxygen species (ROS) to the semiquinone produces an electronically excited state of the resulting quinone, on relaxation producing weak chemiluminescence close to the spectral range of mushroom bioluminescence in the case of unsubstituted quinone [56] . Substituents on the semiquinone influence the electron density on the ring and would dramatically modify the redox potential of intermediates [57] and, therefore, the amount of energy that could be released on relaxation to the ground state. ROS generated as a result of oxygen-dependent redox cycling between dihydroxy and quinone intemediates during melanin formation [58] could be the source of oxidizing reactive species to produce an excited state of a quinolic luciferin, or ROS could be generated by exposure of melanin and precursors to UVR at the cell surface [59] .
Among luciferin precursors that have been proposed, Nakamura in a personal communication cited by Shimomura [12] found that precursors found in Mycena citricolor probably possess a substituted ο-hydroquinone structure. Recently, Purtov et al. [25] provided evidence that hispidine was the precursor of fungal luciferin. Hispidine (4-O-β-d-glucopyranoside) is a naturally occurring styrylpyrone fungal metabolite possessing an ο-hydroquinone moiety as predicted by Nakamura. The potential ability of MnSOD to form a stable semiqui- nonato complex with ο-hydroquinones, together with the slow dismutase mechanism of the “inhibited complex”, in which ROS may interact directly with MnSOD-ligated substrate, suggest possible mechanisms for MnSOD to be directly involved in fungal bioluminescence. We propose that the precursors of mushroom luciferins are catechols (ο-hydroquinones) and that the luciferin(s) in Panellus are semiquinones, which are the anion-radical products of the one-electron oxidation of the substituted hydroquinone.
The following are the part-reactions we propose for luminescence in fungi:
1) one-electron oxidation of a substituted hydroquinone (Q) to produce the semiquinone anion radical in complex with Mn-SOD:
Q + Mn3+ → Mn2+-Q∙−
Purtov et al. [25] provided evidence that 3-hydroxyhispidine, which has a ο-hydroquinone moiety, could be the fungal luciferin. Semiquinones would be produced by oxidation of the ο-hydroquinone moiety by the Mn3+ anion in complex with Mn-SOD, similarly as was observed for Cu/ZnSOD [33] . Soulère et al. [60] observed that a series of catechol derivatives interrupted the dismutation activity of FeSOD, which correlated with the oxidation of the catechols to their ο-quinone derivatives in an irreversible process.
2) non-enzymatic, one-electron oxidation of the Mn-SOD complexed semiquinone radical by a reactive oxygen species such as a hydroxyl radical to produce the excited state of the semiquinone luciferin (Q*):
Q∙− + OH∙ → Q* + OH−
The resulting semiquinone radicals would be oxidized by OH∙ yielding the quinone product Q* in an electronically exited state [56] . Hydroxyl radicals can result from the peroxidatic oxidation of benzoquinone [56] or hydroquinone [55] [61] products of lignolysis.
3) relaxation of the peroxidized (excited) state of the luciferin to produce energy with resultant light emission:
Q* → Q + hν
Generation of the electronically exited state during the reaction of p-benzoquinone with OH• (H2O2) was assumed by Brunmark and Cadenas [56] . The relaxation of that excited state was accompanied by photoemission at 485 - 530 nm, the same spectral range as luminescence in mushrooms. Purtov et al. [25] determined that the in vitro spectrum of luminescence produced using the hispidine luciferin precursor in reaction with cold extract and NADPH was identical to in vivo bioluminesence produced by Mycena chlorophos. Extracellular MnSOD associated with the cytoplasmic membrane or cell wall would be active in the cold extract of these authors.
Shimomura [11] [15] [62] reported observations that appeared to implicate the superoxide anion in the process of mushroom luminescence. However, Shimomura [11] found that Cu/ZnSOD from bovine erythrocytes, conversely to soliciting luminescence as we hypothesize for MnSOD, strongly inhibits the luminescence reaction. Significantly, there are mechanistic differences between Cu/ZnSOD and MnSOD that affect their activities. The dismutase mechanism of human MnSOD appears to differ from other SODs in that the reduced (Mn+2) form reacts with superoxide in two concomitant pathways involving a “burst phase” and a slower “inhibited complex” [51] [52] [63] , the latter evidently formed from an enzyme-substrate complex rather than by superoxide reacting directly with Mn2+ [64] . The “inhibited phase”, allowing interaction of reactive oxygen species with the enzyme-substrate complex, could be associated with bioluminescence. Cu/ZnSOD, by removing superoxide, eliminates ROS which could participate in the luminescence reaction. In addition, Cu/ZnSOD increases the rate of formation of quinones which are reduced by superoxide in a reversible reaction. Quinones themselves are catalysts that increase the rate of autoxidation of hydroquinones [57] removing potential hydroquinone precursors of fungal luciferin. Cu/ZnSOD, unlike MnSOD, has peroxidase-like activities [48] [65] and may oxidize hydroquinones beyond the semiquinone stage to quinone end products [33] . Cu/ZnSOD occurs predominantly in the cytosol in eukaryotes [58] whereas extracellular MnSOD is associated with the cell wall and thus compartmentalized from other SODs [65] . This would explain why luminescence is extinguished when tissues of luminescent mushrooms are disrupted thereby destructing compartmentalization [16] .
Hydroquinones may act as shuttles, moving protons from the inner surface of the cytoplasmic membrane to the outer surface, similar to the action of 2,4-dinitrophenol in mitochondria. The H+ is transported to the outer surface where the hydroquinone is oxidized and the electrons are passed back to metalloproteins. Excess hydroxide ions from the luminescence reaction may accept protons producing H2O, and at the same time quinones/ hydroquinones can be condensed or polymerised outside of the cell forming quinolic polymers or quinhydrones. The participation of hydroquinones in luminescence as luciferin precursors may explain the correlation of mushroom luminescence with ligninolytic fungi [3] . Ligninolysis generates quinolic products which participate in luminescence as luciferin precursers and which are expelled from tissues, oxidized by phenyloxidases and condensed or polymerized as dark- or brown-colored products seen in the growth medium, exudates and cold extracts of luminescent fungal strains. Therefore, luminescence in fungi may provide an evolutionary advantage as an effective mechanism to reduce oxidative stress [66] while simultaneously eliminating excess and potentially toxic quinolic products of lignolysis.
5. Summary
1) The literature on mushroom bioluminescence is reviewed supporting the following, alternate hypotheses for the bioluminescence mechanism in fungi: i) luminescence occurs in the absence of a specialized enzyme and involves the oxidation of an energized state of the fungal luciferin by a reactive oxygen species such as the superoxide anion; or ii) bioluminescence is mediated by a specialized enzyme (the luciferase) acting on the luciferin light-emitter.
2) Differential transcript levels for proteins responsible for post-translational modification (PTM) of enzymes were observed in three separate experiments involving suppression subtractive hybridizations (SSHs) contrasting bioluminescent and non-luminescent strains of Panellus stipticus. Notably, differential transcript levels were observed in all three experiments for proteins involved in ubiquitination which can be determinants for cell localization of proteins or for degradation of proteins in the proteasome.
3) SSH and sequence analysis evidenced that an extracellular isoform of MnSOD was observed in a luminous North American dikaryon isolate of Panellus stipticus which was not observed in a nonluminous European isolate.
4) A hypothetical mechanism for mushroom bioluminescence was proposed whereby the o-hydroquinone moiety of a hispidine derivative ligates with an excreted isoform of MnSOD producing a semiquinone radical complex which reacts with molecular oxygen or other reactive oxygen species producing an excited intermediate which produces light on relaxation. This hypothesis takes into consideration previously published research indicating that mushroom luminescence occurs on mushroom surface tissues [32] , that a catechol derivative is the putative fungal luciferin [25] and that catechols may form stable ligates with SODs [33] .
Acknowledgements
The senior author especially thanks N.S. Manukovskii for many hours of discussions on the biochemistry of mushrooms and possible mechanisms for luminescence.
NOTES
![]()
*Corresponding author.