The Distribution and Expression of BAMBI in Breast Cancer Cell Lines ()

Subject Areas: Pathology
1. Introduction
Breast cancer is one of the biggest hazards to human health. In China, the incidence of breast cancer showed a rapid upward trend in the last two decades. This type of cancer is the leading cause of death in women aged between 35 and 64 years. It has become the primary factor for death caused by cancer in urban female population, which is highly important for researching breast cancer-related genes and investigating the pathogenesis, diagnostic criteria and treatment of breast cancer. The high mortality rate of this type of cancer is attributed to its high capacity to metastasize, mainly to the lungs, liver and bones [1] - [3] .
Breast carcinomas are characterized by their biological complexity and heterogeneity. Initiation and progression of breast cancer is a multi-step process that involves the dysregulation of multiple genes controlling cell survival and proliferation [4] and [5] . BAMBI (BMP and activin membrane-bound inhibitor) gene is discovered by Onichtchouk [6] in 1999, which is located in the area p12.3 - 11.2 in chromosome 10, and codes product as a membrane-spanning glycoprotein of 260 amino acids. Its molecular weight is 29,108 Dalton. It is also known as pseudo receptor, NMA (non-metastatic gene A protein) and belongs to BAMBI family. The family members have a similar structure with the extracellular domain of transforming growth factor―ßI receptor. Therefore BAMBI can be integrated into the ligand-receptor complex and form a polymer with TGF-βII receptor. But because it does not have the serine/threonine kinase domain specific for TGF-βI receptor, it cannot phosphorylate Smad proteins within the cytoplasm, thereby blocking transduction of TGF-β signal at receptor level.
Studies have shown that for breast cancer and other tumors whose origin is epithelial tissue, TGF-β can inhibit the proliferation of tumor cells, and the role of BAMBI negative regulatory mechanism in TGF-β signaling pathway in lung cancer is unclear. In addition, BAMBI reportedly in human lung cancer, colon cancer, liver cancer is present up regulated [7] , but the expression in breast cancer has not been reported.
This study tries to examine the distribution of BAMBI in breast cancer, and explored the relationship between their expression and clinical pathological type and invasive potential using immunofluorescence, Western blot and RT-PCR method. The results showed that according to different pathological type of breast cancer, the stronger the proliferation and invasive ability, the higher the expression of BAMBI.
2. Materials and Methods
2.1. Materials
a. Samples
Three Breast cancer cell lines are MCF-7, MDA-MB-231, MDA-MB-435S which are from Department of Pathology of China Medical University. The cell lines are sort in accordance with the proliferation and migration from high to low (MDA-MB-435s > MDA-MB-231 > MCF-7).
b. Reagents
BAMBI first antibody is goat anti-human polyclonal antibody (molecular weight 29 KD), which was purchased from R & D company; Goat anti-rabbit IgG labeled by Horseradish peroxidase were purchased from Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.; DAB reagent was purchased from MaiXin Biological technology co. LTD in FuZhou. Rabbit polyclonal anti-human ß-actin antibodies (molecular weight 43KD) were purchased from SANTA CRUZ company. Horseradish peroxidase-labeled goat anti-rabbit IgG (H + L), FITC-labeled goat anti-rabbit IgG and PI were purchased from Beijing Zhongshan Golden Bridge biotechnology Co. Ltd. The paraformaldehyde, PBS, H2O2, SDS-PAGE and 2% agarose were got from Institute of Pathology and Pathophysiology in China Medical University.
c. Instruments
The model number of Cell incubator is HeraCell 150 made by Kendro of German. The Polymerase Chain Reaction is ABI9700 made in USA. The Small vertical electrophoresis apparatus is MP3 made by Bio-Rad of USA. The Centrifuge is ST-21 made by SORVALL/KENDRO of USA. The Homogenizer is DIAX100 manufactured by Heidolph of German. The oscillating water bath is GFL made by THERMOLAB of German. The UV spectrophotometer is UV-310 made by PYE-UNIC/SPECTRONIC of UK. The automated electrophoresis gel image analyzer is Chemilmager 5500 made by ALPHA INNOTECH of USA. The microscopic image analysis system is AX70/Coolsnap fx/MetaMorph made by Olympus/UIC of Japan/USA. The laser scanning confocal microscope is TCS SP2 made by Leica of German.
2.2. Experimental Method
a. cell culture
Three cell lines saved in the Department of Pathology of China Medical University were cultured in high glucose DMEM medium containing 10% fetal bovine serum. All cells were incubated at 37˚C, 5% CO2 conditions.
b. Immunofluorescence staining
Cells in logarithmic phase were seeded into the sheet with the growth time more than 24 hours, then were washed three times with 1XPBS, and were in 4% paraformaldehyde for 10 - 20 min. Endogenous peroxidase was blocked within 3% H2O2. Antigen was blocked in 5% bovine serum albumin for 2 h in a humidified box. Primary antibody was added into it, and was incubutaed for the night at 4˚C. Next day, the sheet was washed using phosphate buffered saline (PBS), then incubated in FITC-labeled secondary antibodies at room temperature 1 - 2 h. Cell nuclear was couterstained by PI in 10 min after being washed by PBS, then was mounted after being washed again. We observe it using laser scanning confocal microscope system, and get the clearest figure through the Z-axis changes.
c. Western blot
Cells were centrifuged at 810 g/min for 5 min at 4˚C. An appropriate amount of lysis buffer and protease inhibitors added into the collected cells. After ultrasonic decomposition, we got the supernatant through centrifuging (4˚C 12,000 g/min) for 15 min. Determination of protein content was done with Coomassie brilliant blue. The mixture of supernatant, ultra-pure water and loading buffer in appropriate proportion were thoroughly shaken, and in boiling water bath for 5min.We took 10 μL sample (containing 30 μg of total protein) and put it onto 5% spacer gel 80 V for 15 min, then 12% separating gel 120 V for 80 min. After the electrophoresis in sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE), the protein was transferred to PVDF membrane by transferring in 50 V for 1.5 h.
The membrane was removed, and mounted in 5% skimmed milk. The membrane was put into the primary antibody (1:1000 dilution), incubated at 4˚C for the night. The next day secondary antibody (1:10,000 dilution) were added and incubated at room temperature for 2 h. Then we took the membrance exposure in darkroom in ECL reagent and captured images using automated electrophoresis gel image analysis system after developing fixed and measured strip integrated optical density. With, with BAMBI/β-actin ratio as a relative expression level of BAMBI, we repeated this experiment which took β-actin as internal control and BAMBI/β-actin ratio as a relative expression level of BAMBI, and averaged.
d. RT-PCR
RNAout was used to extract total RNA, according to instructions. The total RNA was measured by UV spectrophotometer A260/A280, the ratio is 1.9 to 2.1. BAMBI primers: the Upstream BAMBI-F: 5-GCACCACCATACCCACATTG-3, downstream: BAMBI-R: 5-GCAGCCTCTTATTTTCACTTCG-3. The fragment length is 282 bp, Tm: 59/59. GAPDH primer: upstream GAPDH-F: 5-GAAGGTCGGAGTCAACGGAT-3, downstream GAPDH-R: 5-CCTGGAAGATGGTGATGGG-3. The fragment length is 224 bp, Tm: 58.7/57.9. Oligo dT-Adaptor Primer was used to synthesize the first strand of cDNA synthesis. 1 μL cDNA taken from each sample was amplified using PCR. The reaction happened in 25 μL reaction system, and the conditions were denaturation in 94˚C for 5 min, the appropriate number of cycles 94˚C 30 s, 64.1˚C 30 s, 72˚C 45 s, and finally extending in 72˚C for 10 min. The amplification products of BAMBI, GAPDH were collected images by automated electrophoresis gel image analysis system after the gel electrophoresis with 2% agarose and determined the strip integral optical density, which were using 100 bp DNA Marker as molecular weight standards. In order to get the relative contents of BAMBI, we consulted the value in GAPDH to standardize the one of BAMBI. This experiment was repeated and gets the averaging.
2.3. Statistical Analysis
The statistical analysis for all data was made through SPSS 12.0 software. Measurement data were expressed as mean ± standard deviation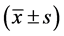 ; The variance (ANVOA) was used as the comparative analysis for means of more than two groups, and the significant difference was 95%.
; The variance (ANVOA) was used as the comparative analysis for means of more than two groups, and the significant difference was 95%.
3. Results
3.1. Immunofluorescence Staining Cells
Previous study suggested that [8] , immunofluorescence showed that BAMBI was located in both nucleus and cytoplasm in porcine granulosa cells. That is different with the location of BAMBI in the cell lines of breast cancer, but both of them showed the up-regulation of BAMBI.
![]() (a)
(a)![]() (b) (c)
(b) (c)
Figure 1. Immunofluorescence assay on the expression of BAMBI protein in breast cancer cell line (a) MDA-MB-435S; (b) MDA-MB-231and (c) MCF-7.
3.2. Western Blot Result
Three strains of cellular proteins at 29 KD showed clear bands that are consistent with the molecular weight of BAMBI. We capture images by automatic electrophoresis gel image analysis system, which is shown in Figure 2 and the comparison of BAMBI protein expression in three cell lines are given in Table 1.
The order of BAMBI protein expression in the three breast cancer lines is MDA-MB-435s > MDA-MB-231 > MCF-7. Respectively the difference was significant (P < 0.05), which is corresponding to BAMBI mRNA expression of the three cell lines, and data from related reference [9] shows over expression of BAMBI mRNA in an in vitro model led to significantly increased proliferation and migration (+581.2%, P = 0.004). The Western blotting results is consistent with the hypothesis of this study.
3.3. RT-PCR Result
The amplification products of the three cell lines by electrophoresis presented bands whose fragment length is 282 bp (Figure 3), which is consistent with the fragment length designed in primer. Captured images automatically through electrophoresis gel imaging analysis system, as shown below;
The order of BAMBI mRNA expression in the three breast cancer cell lines is MDA-MB-435s > MDA-MB- 231 > MCF-7. Respectively the difference was significant (P < 0.05), which is corresponding to the proliferation and migration of the three cell lines. Data from related reference [9] shows over expression of BAMBI mRNA led to significantly increased proliferation and migration (+581.2%, P = 0.004), which is accordant to ours (Table 2).
Relative molecular
Weight MDA-MB-435S MDA-MB-231 MCF-7
![]() BAMBI
BAMBI![]() β-actin
β-actin
Figure 2. Western blotting detects BAMBI protein expression in three breast cancer cell lines.
bp M 1 2 3 4 5 6
![]()
Figure 3. BAMBI mRNA expression in three strains of cell lines. Label: 1 = BAMBI (MCF-7); 2 = BAMBI (MDA-MB-231); 3 = BAMBI (MDA-MB-435 s); 4 = GAPDH * 7; 5 = GAPDH * 231; 6 = GAPDH * 435 s.
![]()
Table 1. The compare of BAMBI protein expression in three cell lines.
Label: *P vs MDA-MB-435S; **P vs MDA-MB-231; ***P vs MCF-7.
![]()
Table 2. The compare of BAMBI mRNA expression in three cell lines.
Label: *P vs MDA-MB-435S; **P vs MDA-MB-231; ***P vs MCF-7.
4. Discussion
In this study, BAMBI was found expressed in the cell membrane and cytoplasm close to the membrane of breast cancer. This has been confirmed by use two different BAMBI antibodies for Immunofluorescence. Three breast cell lines expressed BAMBI protein and mRNA in the order of expression quantity which consistent with their proliferation and migration, which was confirmed by use Western Blot and RT-PCR. BAMBI was also detected in ovarian cancer as has been already shown [9] . This may indicate that BAMBI has distinct functions in proliferation and migration of breast cancer.
BAMBI is a similar gene in the human genome and has strong homology in sequence as Xenopus BAMBI gene. In fact, many of the features of human gene BAMBI are obtained from the study of Xenopus BAMBI gene.
In Xenopus’s embryos, Onichtchouk et al found pseudo receptor BAMBI whose structure is similar to TGF- βI receptor which can combine with TGF-βII type receptors competitivly, induce ligands-TGF-β superfamily (TGF-βs, BMPs, activins like) to form the ligand-receptor complex. Under the effect of BMP ligand , Xenopus embryo expresses BAMBI, and effects on type I or type II receptor stably which thereby makes them lose the ability to combine with TGF-β.
Due to the lack of serine/threonine kinase domain which is in the intracellular area of type I receptor, BAMBI doesn’t have the serine/threonine kinase activity, and cannot phosphorylate SMAD protein in cytoplasmic domain. Thereby TGF-β signal is blocked and a series of downstream genes cannot express well.
Additionally, BAMBI can form homodimmer with type I receptors, which inhibites the formation of receptor complex and make TGF-β signal response into a static state. The antagonism is induced by the BMPs and is similar to negative feedback regulation. Some previous study detected BAMBI’s expression on animals and verified results supported this study [10] - [14] .
As the structural features of BAMBI, it is extensively involved in the regulation of TGF-β signal, which plays an important role in the occurrence of tumors that originate from epithelial tissue.
Sekiya et al. [7] reportedly have detected that the BAMBI expresses high abnormally in human colon cancer and liver cancer in 2004, using immunohistochemistry, semi-quantitative RT-PCR, but low in the corresponding adjacent normal tissues.
At the same time, they proved that there was a binding site in BAMBI promoter which is corresponding in BMP. Therefore its transcription is regulated by BMP. And they confirmed that BAMBI is a downstream gene in β-catenin/TCF-4 signal transduction pathways. Thereby we inferred that Cancer occurs due to the transcriptional of BAMBI activated by β-catenin and the negative feedback regulation of BAMBI to TGF-β signal causes the cancer cells to escape from the growth inhibition of TGF-β.
Sekiya et al. [15] did research on NMA in MKN28 and other three gastric cancer cell lines and found the negative feedback regulation of NMA to TGF-β signal which causes the cells to escape from the growth-inhibi- tion mechanism mediated by TGF-β and induces gastric cancer cell’s growth and invasion.
Studies have shown that, TGF-β signal normally inhibited proliferation of epithelial cells, and its down regulation or loss of function may cause cancer. And along with the growth of tumor, TGF-β signal is closely related to the development, angiogenesis and metastasis of tumor.
BAMBI and TGF-βI receptor combine with ligands competitively. Therefore the upregulation of BAMBI can block TGF-β more. In addition, BAMBI acts as the cause of type I and type II receptor down regulation or loss of functionality. Although the thesis of BAMBI as a pseudoreceptor is well received, it may also be explained by modulation of TGF-β signaling in nucleus, leading to the observed pro-oncogenic properties.
In summary, these findings provide new insights into the relationship between BAMBI and the proliferation and metastasis of breast cancer, which verifies our indication at the molecular level.
5. Conclusion
The results showed that the positive reaction of BAMBI was observed in membrane and cytoplasm close to membrane of breast cancer. Our findings suggest the expression level of BAMBI’s protein and mRNA in breast cancer cells is relevant to breast cancer’s ability to proliferate and metastasize and increases with the proliferation and metastasis strengthened. The correlation indicates that BAMBI can be used as a biomarker for clinical stages of breast cancer to guide the clinical treatment for it.
NOTES
![]()
*Corresponding author.