Construction of EST-SSR Databases for Effective Cultivar Identification and Their Applicability to Complement for Lettuce (Lactuca sativa L.) Distinctness Test ()
1. Introduction
Lettuce (Lactuca sativa L., Asteraceae) is one of the most widely cultivated vegetables [1] . Sixty-three lettuce cultivars obtained Plant Variety Protection (PVP) at the Korea Seed & Variety Service. This is the fourth highest number among vegetable crops in Korea (http://www.seed.go.kr). A new plant variety requires novelty, denomination, distinctness, uniformity, and stability (DUS) to get registered for plant breeder rights based on the International Union for the Protection of New Varieties of Plants (UPOV) regulations. The morphological characters are used for the DUS test. It is necessary to conduct two growing cycles at the fields or greenhouse for growth trials. Moreover, morphological characters are quantitatively inherited and their expression is affected by environmental factors. Thus, a rapid and robust “DNA marker technique” has been used to identify cultivars for the DUS test [2] - [4] .
DNA markers have many advantages to identify cultivars due to their independence from environmental influences. The UPOV suggests that simple sequence repeat (SSR) markers are suitable for a DNA profiling database due to their multi-allelic nature, reproducibility, high polymorphism, easy automation, and co-dominant inheritance [5] . SSRs are present in both coding and noncoding regions [6] . Expressed sequence tags (ESTs)- SSRs have advantages over genomic SSRs representing noncoding regions because they are present in coding regions and the expressed sequence data can be easily retrieved from the public databases [7] . Moreover, EST- SSRs may enhance the applicability of DNA markers by expressing the variation in transcribed genes [8] . How- ever, the development of SSR markers in lettuce has been very limited because it is costly, and labor intensive. There have been above 300 SSR markers described in the literature [9] - [11] . In addition, there are no reports on applicability for the DUS test using molecular marker in commercial lettuce cultivar. Therefore, we constructed EST-SSR profiles databases to effectively identify 92 cultivars widely cultivated in Korea and investigated the possibility of utilization in the distinctiveness tests by comparing morphological characterization of lettuce cultivars with limited variability through SSR analysis.
2. Materials and Methods
2.1. Plant Materials and DNA Extraction
92 commercial lettuce cultivars were used in this study (Table 1). DNA was extracted from the 20 seeds of each cultivar. DNA isolation was carried out using NucleoSpin® Plant II (Macherey-Nagel Cat. 740 770.250) based on the manufacturer’s protocol. DNA concentration was adjusted to 20 ng using Nanodrop (Thermo Scientific, Rockford, IL, USA) and then used for polymerase chain reaction (PCR) analysis.
2.2. Screening of the Lettuce EST Database and Primer Design
A total of 81,330 ESTs were downloaded from NCBI (to July 1, 2011) and assembled with CAP3 [12] . A web tool called microsatellite identification (MISA) (http://pgrc.ipk-gatersleben.de/misa/) was used to search for SSRs in the unigenes with a minimum of six repeats units for a di-nucleotide, five repeats units for a tri-nucleo- tide, five repeats units for a tetra-nucleotide, four repeats units for a penta-nucleotide and hexa-nucleotide. SSR primers were designed with the Primer 3 program [13] and were used to generate PCR products of 100 - 300 bp at annealing temperatures (Ta) of 50˚C - 60˚C. Primers were synthesized by Bioneer Company (Daejeon, Korea).
2.3. PCR and Electrophoresis
The 370 SSR primers were used to identify lettuce cultivars. PCR amplifications were performed in 25 µL volume containing 20 ng template DNA, 10 pmol∙µL−1 forward and reverse primers, 2.5 mM dNTP mixture, 10× PCR buffer solution, and 1 unit Taq polymerase (Genet Bio, Seoul, Korea). All primer combinations consisted of a 4 min initial denaturation at 94˚C followed by 40 PCR cycles of 30 s at 94˚C, 30 s at 55˚C, 45 s at 72˚C, and a final 10 min extension at 72˚C on T professionalTM thermocycler (Biometra, Göttingen, Germany). To select informative SSR markers, seven cultivars (“Gangpungjeokchima”, “Numberone”, “Icered”, “Eboniblack”, “Topgreen”, “Chirivel”, and “Ace”) among the 92 cultivars were first screened to select polymorphic SSR markers. The PCR products of the seven cultivars were separated on denatured 6% polyacrylamide gels and then silver stained with the DNA Silver Staining Kit (Promega Cat. Q4132, Madison, WI, USA) according to the manufacturer’s instructions. From the genotyping result of the seven lettuce cultivars, SSR primer pairs with
![]()
Table 1. The 92 cultivars used to construct the DNA profile database using EST-SSR markers.
highly reproducible and clear band patterns were respectively labeled at the end site of the forward primer with FAM, VIC, NED, and PET dye and then PCR amplification was performed. The PCR products (4 µL) were separated on 2% agarose gels and then 1 - 3 µL was mixed with 200 µL water depending on the intensity of the PCR products. 1.5 µL aliquot of diluted PCR product, 10 µL of deionized formamide and 0.25 µL of size marker (LIZ500 size standard) were mixed and denatured for 5 min at 94˚C. The PCR products of 94 cultivars were analyzed by capillary electrophoresis (Genetic Analyzer 3130XL, Applied Biosystems, Foster City, CA, USA) using the manufacturer’s instructions. The allele determination for the SSR markers was evaluated with the GeneMapper 3.7 software program (Applied Biosystems).
2.4. Data Analysis
Peaks were scored with regard to the presence and absence of peaks for genotypes. Scores of “1” and “0” designated the presence and absence of peaks for each SSR marker allele, respectively. Polymorphism information content (PIC) was calculated via the formula established by [14] :
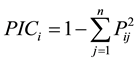
where Pij is the frequency of the jth allele for the ith locus summed across all alleles for the locus. The cluster analysis was based on Jaccard’s similarity coefficient using the unweighted pair group method using arithmetical averages [15] method in the NTSYSpc 2.10b [16] software package. Mantel test was employed for the correlation between genetic and morphological distances [17] .
2.5. Morphological Analysis
Morphological characters were analyzed according to the DUS characteristics of the lettuce test guideline prescribed by the Korea Seed & Variety Service (http://www.seed.go.kr). 29 were used among 39 morphological characters of lettuce for the DUS test.
3. Results and Discussion
3.1. Development of EST-SSR Markers
With 81,330 lettuce ESTs from the NCBI database, 41,609 singletons and 8452 contigs were identified by CAP3 software. In total, 4229 SSR loci from singletons [11] and 807 SSR loci from contigs were identified using the MISA program. Of 807 SSR loci, the highest proportion was trinucleotides (455, 56.4%), dinucleotides (266, 33%), hexanucleotides (42, 5.2%), pentanucleotides (31, 3.8%), and tetranucleotides (13, 1.6%). These results support the previous studies that trinucleotide repeats occurred in higher frequency than different repeat motif in lettuce [7] [18] .
3.2. EST-SSR Polymorphisms in Lettuce
The 348 SSR primers developed previously [9] -[11] and 22 SSR primers among 710 primers designed in this study were used to screen in seven cultivars. Of the 370 SSR primers, 82 showed polymorphisms among tested cultivars (Table 2). 58 of the 82 polymorphic markers were selected on the basis of easy scoring, high reproducibility and peak quality among cultivars. Thereafter, 92 cultivars were examined using 58 SSRs by an automated DNA sequencing system (Figure 1). The PIC, number, and size of the alleles for the 92 cultivars were analyzed using the 58 EST-SSR markers (Table 3). A total of 176 alleles were obtained ranging from 2 - 8 alleles with an average of 3.03 alleles. The polymorphism information content (PIC) values, which are reflective of allele diversity and frequency among the varieties, were not uniformly high for the tested SSR l˚Ci. The average PIC value was 0.425 and it ranged from 0.022 (SML-052) to 0.743 (KSL-26). The variation at SSR loci in 92 cultivars are summarized in Table 3.
Several reports have described PIC value of SSR analysis in lettuce. This result was higher than 0.32 reported by [10] but lower than 0.55 of [19] and 0.56 of [20] . This may be mainly due to genetic background of tested cultivars and chromosome loci linked to SSR marker. Generally, the level of polymorphism for the EST-SSR
![]()
Table 2. SSR markers screened to identify lettuce cultivars and polymorphism of the SSR markers.
![]()
Figure 1. Amplified SSR fragments of 3 lettuce cultivars at SML-022 locus. The PCR products were separated using a Genetic Analyzer 3130XL (Applied Biosystems, USA) and detected using GeneMapper software (Applied Biosystems, USA). Lane 1: “Joara”, Lane 2: “Daepungjeokchima”, Lane 3: “Peoseutgyeolgu”.
![]()
![]()
Table 3. Description of 58 polymorphic EST-SSR markers selected to construct a DNA profiling database for lettuce.
markers was lower than that for the genomic SSR markers. The reason might be that EST-SSR markers are based on the translated region [21] . However, the set of 58 EST-SSR markers selected in this study might be very informative for identifying 92 commercial lettuce cultivars in Korea.
3.3. Genetic Relationships among the Lettuce Cultivars
A dendrogram based on the similarity coefficients of the 92 cultivars was constructed. The dendrogram scale varied from 0.29 to 1.00 (Figure 2) and were clustered into four major groups at a similarity level of 0.40. Cluster I consisted of cultivars with red leaf lettuce, which was split into three clusters with a mean similarity of 0.47.
![]()
Figure 2. Phenetic dendrogram of 92 lettuce cultivars based on SSR markers. The scale at the bottom is Jaccard’s coefficient of similarity. The numbers (1 - 92) at right refer to the list of cultivars in Table 1.
Cluster II consisted of cultivars with Crisphead, Romaine, Green leaf, and Butterhead, which split into two clusters with a mean similarity of 0.43. Cluster II-1 consisted of cultivars with Crisphead, Romaine, Green leaf, and cluster II-2 cultivars with consisted of the Butterhead type. Crisphead was clustered in one group with a similarity level of 0.70. Cluster III consisted of cultivars with green leaf and black leaf at a similarity level of 0.47. Cluster III-1 contained cultivars with green leaf, and cluster III-2 contained black leaf except for “Cheongpungchima” and “Greencheongchima”. Cluster IV contained cultivars with the green leaf of “Greenglace” and “Scanstar”. Generally, the clustering of 92 lettuce cultivars was mainly categorized into 4 major groups corresponding to morphological traits. However, eight cultivars (Group 1: “Cheongpungchima” and “Greencheongchima”, Group 2: “Sambokmeokchima”, “Heukssammeokchima”, and “Meokdoli”, Group 3: “Yeoreumgohyangdambae”, “Cheonpung”, and “Sijeoseugreen”) were not discriminated by the 58 EST-SSR markers. These cultivars may be developed by parents with narrow genetic background, and a limited number of SSR markers were used to identify for lettuce cultivars.
3.4. Correlation between EST-SSR Markers and Morphological Characters
We investigated morphological traits for 3 pairs with 100% genetic similarity. However, cultivars in each pairs were not distinct for the 29 morphological traits under same environmental condition (Table 4 and Figure 3). In
![]()
Figure 3. Morphological description of the eight cultivars representing 100% genetic similarity. The numbers in the pictures represent lettuce cultivar numbers in Table 1.
![]()
![]()
Table 4. Some morphological characteristics of the eight cultivars used for the distinctness test.
QL: qualitative, QN: quantitative, PQ: pseudo-qualitative. The upper numbers refer to the list of cultivars in Table 1.
addition, lettuce breeders were in agreement with our results of morphological traits. The correlation between molecular and morphological data for 8 cultivars in 3 pairs was analyzed by the [17] and revealed a good fit between the two data (r = 0.82). These results may be due to using large number of SSR markers derived from EST representing coding regions, and precise genotyping through DNA sequencing system for cultivar identification.
UPOV established the possible use of molecular markers for a DUS test [22] . They suggested four possible application models by technical committee based on research by the working group on Biochemical and Molecular Techniques, and DNA-Profiling in Particular (BMT). Option 1 is to use molecular markers that are directly linked to traditional characteristics (gene-specific markers). Option 2 is to calibrate the threshold levels for molecular characteristics against the minimum distance for traditional characteristics. Option 3 is to use the molecular marker characteristics. Option 4 is combining phenotypic and molecular distances to manage a cultivar collection. Of the four options, options 1, 2, and 4 are positive assessment models. Option 3 is not a positive assessment [22] . We tested the morphological characteristics for the cultivars with 100% genetic similarity on the basis of molecular markers. Their correlation was a good fit (r = 0.82). Thus, we conclude that the EST-SSR markers selected in this study will be useful for choosing the most similar cultivars to candidate cultivars, protection of plant breeder’s rights, and an alternative choice to conduct a DUS test in lettuce.
NOTES
*Corresponding author.