Modeling Metal Binding Sites in Proteins by Quantum Chemical Calculations (Short Report)
In protein binding sites Mg2+ is usually bound to the side chains of Asp/Glu or Asn/Gln, or the backbone peptide group [5] . Accordingly, the side chains of Asp/Glu, Asn/Gln, and peptide backbone group are modeled as acetate (CH3COO−), acetamide (CH3CONH2), and N-methylacetamide (CH3CONHCH3), respectively. In proteins as in aqueous solution Mg2+ is mostly hexacoordinated [6] [7] . Thus, Mg2+ complexes are modeled as MgL6 (L = H2O, CH3CONH2, CH3CONHCH3, CH3COO−). The most common coordination number of Li+ is four [8] . Accordingly, its complexes are modeled as LiL4.
The B3LYP/6-31+G(3d, p) method/basis set combination is chosen among various other combinations as it best reproduces the experimental dipole moments, µ, of model ligands and metal―ligand bond distances, R, in metal complexes [9] [10] :
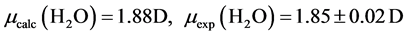
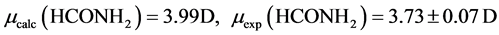
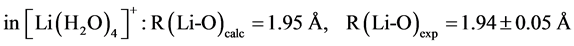 ,
,
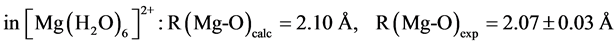 .
.
Condensed-phase calculations are calibrated with respect to experimental free energies of hydration of the metal cations and model ligands. Results are given in Table 1. For validation purposes, the experimental free energies of Li+ → Mg2+ substitution in acetate, oxalate and nitrilotriacetic acid complexes are used and com-
![]()
Table 1. Comparison between computed and experimental hydration free energies,  , of metal cations and model ligands (in kcal/mol) [9] .
, of metal cations and model ligands (in kcal/mol) [9] .
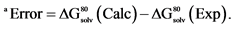
![]()
Table 2. Comparison Between Computed and Experimental Free Energies (in kcal/mol) for [MgX] + [Li(H2O)4]+ → [LiY] + [Mg(H2O)6]2+ in Water, ΔG80 (Li+ → Mg2+) [9] .
aError = ΔG80(Calc) − ΔG80(Exp). bNTA = nitrilotriacetic acid bound in a tetradentate fashion (including central N atom) to the metal.
pared with those evaluated theoretically using the models and methods described above. As seen from the results presented in Table 2, the calculations reproduce the experimentally determined free energies of Li+ → Mg2+ substitution within1 kcal/mol. Now, avenues are open for the “production run”.