Journal of Cancer Therapy
Vol.06 No.04(2015), Article ID:55565,10 pages
10.4236/jct.2015.64037
Efficacy of 5-FU or Oxaliplatin Monotherapy over Combination Therapy in Colorectal Cancer
Maria Toloudi, Panagiotis Apostolou, Ioannis Papasotiriou*
Research Genetic Cancer Centre Ltd. (R.G.C.C. Ltd.), Florina, Greece
Email: *office@rgcc.genlab.com
Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 14 March 2015; accepted 10 April 2015; published 13 April 2015
ABSTRACT
Recent clinical and research development supports the use of 5-fluorouracil in combination with oxaliplatin for the treatment of patients with advanced colorectal cancer (CRC). 5-Fluorouracil (5-FU), which is as an anti-metabolite, is a widely used cytostatic drug. Although the rate of response, quality of life and overall survival differs between CRC patients, the above combination remains a widely used chemotherapeutic regimen. In some cases, a cancer stem cell (CSC) population may resist the majority of chemotherapeutic models. This study investigated if monotherapy is more efficacious than 5-fluorouracil and oxaliplatin combined for the treatment of CRC, using a CRC cell line and a CSC-like line. Cell viability was evaluated by cellular-based assays, and quantitative polymerase chain reaction (q-PCR) assays were performed to assess the expression of specific genes (TYMS, DNMT1, NANOG, DHFR, SHMT1, ERCC1, DPYD) correlated with 5-FU and oxaliplatin resistance. We observed that 5-fluorouracil was more effective in both CRC and CSCs. This find- ing proved the hypothesis that, in some cases, monotherapy may be more successful in CRC treatment than a drug combination that may be cytotoxic and inflict adverse side effects.
Keywords:
5-Fluorouracil, Oxaliplatin, FOLFOX, Colon Cancer Stem Cells

1. Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in worldwide. The lifetime risk of developing CRC is approximately 5%, depending on the individual and other risk factors, and 5-year survival for advanced stage CRC is only 12% [1] . The management of CRC focuses on curative or palliative treatment. The first can be achieved by surgery, chemotherapy or radiation, while the second is applicable in incurable CRC [2] [3] . Among the available chemotherapy drugs, 5-fluorouracil (5-FU) has been the mainstay treatment. However, new small molecule drugs have been recently developed, including irinotecan, oxaliplatin, oral fluoropyrimidines and raltitrexed [4] . One established chemotherapy regimen for the treatment of CRC is FOLFOX, which consists of a combination of leucovorin (FOL), 5-FU (F) and oxaliplatin (OX). According to experimental data, there is controversy concerning whether monotherapy or combination therapy is better [5] [6] . Although chemotherapy kills most cells within a tumor, it has been experimentally demonstrated that is ineffective in cancer stem cells (CSCs), which exhibit resistance to chemotherapy [7] . Several factors have been found to be correlated with the resistance of CSCs to chemotherapy and radiotherapy. According to a study, the Wnt signaling pathway is implicated in stem cell survival [8] . In glioblastoma, it has been demonstrated that the CD133 cells display strong capability on tumor’s resistance to chemotherapy and this resistance is probably due to high expression of BCRP1 and MGMT, as well as anti-apoptosis protein and inhibitors [9] . Another study, aimed that ABC transporters are implicated in drug resistance of CSCs, through the expression of multifunctional efflux trnasportes from the ABC gene family [10] .
The present study aimed to investigate the effect of 5-FU and oxaliplatin in CRC and colon CSCs lines when the abovementioned drugs are used in combination or as monotherapy.
2. Materials and Methods
2.1. Cell Lines and Culture Conditions
A human (parental) colon CSC line (36112-39P) was obtained from Celprogen Inc. (San Pedro, USA). Cells were cultured in 25 cm2 flasks in Human (Parental) Colon Cancer Stem Cell Culture Serum-Free Media (M36112-39P; Celprogen). HCT-116 colon adenocarcinoma cells were obtained from the European Collection of Cell Cultures (ECACC, UK). Cells were cultured in the indicated culture medium with the appropriate amount of heat-inactivated fetal bovine serum (FBS; S0615; Biochrom, UK) and 2 mM L-glutamine (G7513, Sigma, Germany), and were incubated in a humidified incubator at 37˚C with 95% O2 and 5% CO2. Media and supplement changes were performed when 90% confluence was obtained. The cells were obtained by trypsinization with 0.25% trypsin-EDTA (25200-072; Invitrogen, California, USA) during the exponential phase of cell proliferation. All experiments were performed after the cultures reached 80% - 90% confluence.
2.2. Anticancer Agents
5-FU (F6627) was obtained from Sigma Aldrich (St. Louis, MO, USA) and reconstituted in ammonium hydroxide (ΝΗ4ΟΗ) to a final concentration of 50 μM and stored at 4˚C until required. The same procedure was followed for oxaliplatin (O9512; Sigma Aldrich), which was reconstituted in distilled water to a final concentration of 1.12 μM.
2.3. Cell Treatment and Exposure Time
The drug concentrations used in most of the combination studies were based on the IC50 values, which are characterized as the drug concentration responsible for 50% growth inhibition. In this study, the concentrations of drugs used were, as much as possible, relevant to those used in clinical practice. To determine the cytotoxicity of each agent separately, as well as in combination, each cell line was plated in a 24-well plate format and incubated for 24 h at 37˚C. During the exponential growth phase, cells were treated with 50 μM 5-FU and 1.12 μM oxaliplatin for 72 h, 144 h, 192 h and 240 h. The protocol included a cell population treated with 5-FU alone, a second population treated with oxaliplatin alone, and a third population treated with FOLFOX. Untreated cells were used as controls. At the end of each incubation period, cytotoxicity was evaluated using three viability/co- lorimetric assays as well as flow cytometric analysis. The cell number was also determined using the NucleoCounter NC-100 instrument (ChemoMetec A/S, Denmark). Finally, the expression of specific genes was measured.
2.4. Viability/Colorimetric Assays
Methyltetrazolium (MTT), sulforhodamine B (SRB) and crystal violet (CV) colorimetric cytotoxicity assays were used to measure enzymatic activity and quantify the population of living cells using indirect parameters. Cytotoxicity was measured when cells (18.000 cells/well) were seeded into 96-well cell plates in 200 μl culture medium for 24 h prior to drug exposure. Cells were then treated with 5-FU, oxaliplatin, and their combination. After drug exposure, 20 μl of MTT (5 mg/ml) (M2128; Sigma) was added to each well and cells were incubated for 3 h at 37˚C. The supernatant was aspirated and the cells were washed with phosphate buffered saline (PBS; P3813; Sigma Aldrich). Next, 100 μl dimethyl sulfoxide (DMSO) (472301; Sigma) was added and homogenization of the sample was achieved by gently pipetting up and down. At the end of the procedure the optical density (O.D) was measured at 570 nm. An additional wavelength at 630nm was used to remove noise from the external conditions affecting the measurement [11] - [17] . During SRB assays, plated cells were fixed by layering 50 μl of 10% trichloroacetic acid (TCA; 91228; Fluka) and incubated at 4˚C for 1 h. Cells were stained by the addition of 100 μl/well 0.4% SRB (341738; Sigma Aldrich). The unbound stain was removed by washing twice with 1% acetic acid. To release the bound dye, 200 μl of 10 mM Tris buffer pH 10.5 (T6791; Sigma Aldrich) was added and absorbance was measured at 570 nm and 690 nm [18] - [21] . Finally, for CV assays, the medium was aspirated and cells were fixed by adding 100 μl of 10% formalin (1.0400 3.2500; Merck) for 20 min at room temperature. At the end of the fixation procedure, cells were stained by adding 100 μl of 0.25% aqueous crystal violet (HT901; Sigma Aldrich) for 10 min at room temperature. The last step included washing of the plate with distilled water, the homogenization of cells by using 33% glacial acetic acid (401422, Carlo Erba) and measurement of the optical density of living cells at 570 nm and 690 nm [22] - [24] .
2.5. qRT-PCR
Total RNA was extracted from treated and untreated cells using the RNeasy mini kit (74105; QIAGEN). RNA samples were evaluated spectrophotometrically and on agarose gels by checking the 18S-28S bands. Then 1 μg RNA was used as the template for cDNA synthesis using an iScript cDNA synthesis kit (1708891; Bio-Rad). The above strand was used as template for the quantitative real-time (qRT)-PCR reaction (50 ng/reaction), which was performed using iTaq Universal SYBR Green Supermix (1725124; Bio-Rad). Specific primers for each marker and for the endogenous control gene (18S rRNA) were designed with Genamics Expression 1.1 software (Genamics, Hamilton, New Zealand). The sequence of primers was run on BLAST to exclude those that amplified undesired genes (Table 1). The PCR reaction program was as follows: initial denaturation at
Table 1. Primer pairs that were used in qPCR.
95˚C, 50 cycles of denaturation at 95˚C for 10 sec, followed by annealing at 59˚C for 30 sec. A final extension step was performed at 72˚C for 10 min followed by melting curve analysis. Data were analyzed according to the Livak method [25] .
2.6. Cell Number Quantification
To determine the number of viable cells after exposure to the drugs, the Nucleo Counter NC-100 image cytometer was used. This technology is based on detection of fluorescence from DNA-bound fluorescent dye, propidium iodide (PI). The Nucleo Cassette that is compatible with the NC-100 can count even the most aggregated mammalian cells.
2.7. Statistical Analysis
All experiments were performed in triplicate for all assays and each cell line. The statistical significance of the effect of the selected compound was evaluated by the “difference of the means” test. A P value < 0.05 was considered statistically significant.
3. Results
3.1. Cell Lines and Culture Conditions
HCT116 cultures treated with 5-FU exhibited similar confluence with those treated with FOLFOX after 72 h, 144 h and 192 h of drug incubation. Their confluence levels were compared with their respective untreated control cells, which had almost the same proliferation levels as oxaliplatin-treated cells. Growth inhibition was more evident in 5-FU-treated cells after 240 h of treatment (Figure 1). Similar results were observed in colon CSCs, but the effect of FOLFOX seemed to be slightly higher than the other two monotherapies after 144 h, while growth inhibition was much higher in 5-FU-treated cells either after 192 h or 240 h of incubation (Figure 2).
3.2. Viability/Colorimetric Assays
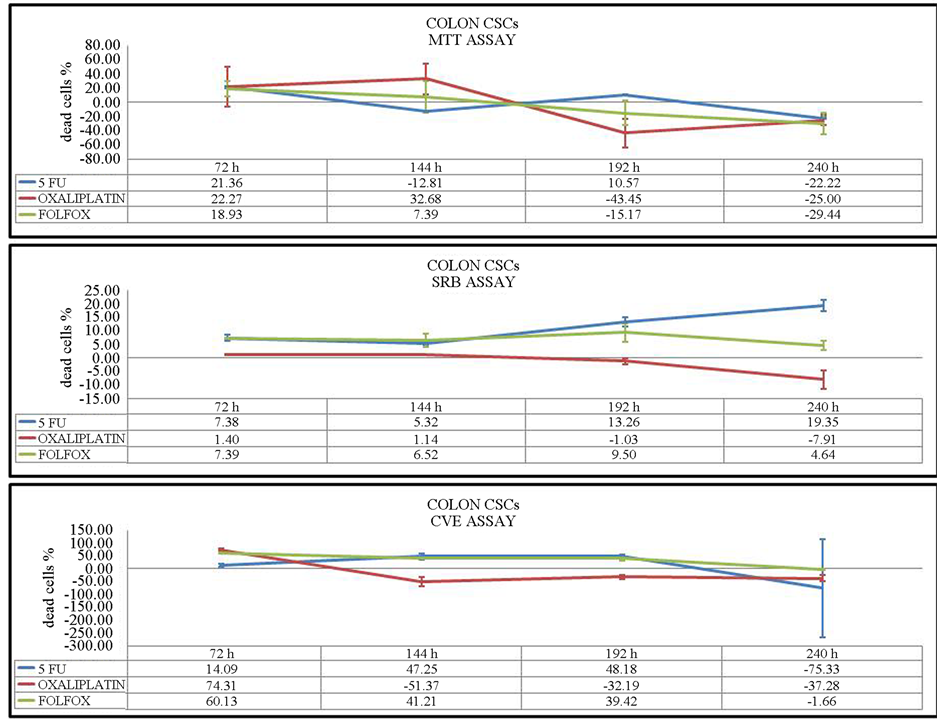
To elucidate the time-dependent effects of 5-FU, oxaliplatin and FOLFOX on colon CSC viability, we first assessed treatment efficacy at 72 h. Oxaliplatin inhibited cell proliferation at a higher level, as measured by MTT
Figure 1. HCT-116 CRC cells pre- and post-treatment with 5-FU, oxaliplatin and their combination over 240 h incubation. Unstimulated indicates cells without the addition of drugs.
Figure 2. Colon CSC stem cells pre- and post-treatment with 5-FU, oxaliplatin and their combination over 240 h incubation. Unstimulated indicate cells without the addition of drugs.
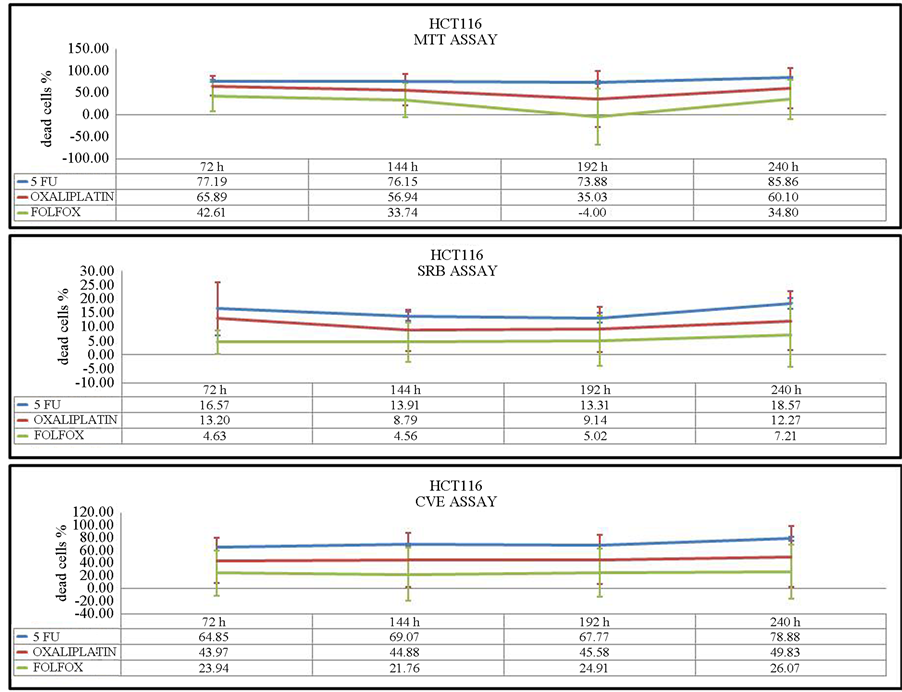
and CV assays. After 144 h of incubation, oxaliplatin as well as 5-FU were more toxic in the MTT and CV assays. SRB assays showed an equal or slightly higher effect of FOLFOX in comparison with 5-FU after 72 h and 144 h of incubation. Concerning the results of the three assays after 192 h and 240 h, the viability of cells was significantly inhibited by 5-FU at both time points (Figure 3), while 5-FU had the greatest effect on HCT116 cells at 144 h, 192 h and 240 h. After 72 h of drug treatment, oxaliplatin inhibited HCT-116 cell viability to the greatest extent. During this incubation time, the toxicity of oxaliplatin was the lowest compared with 5-FU. (Figure 4).
3.3. qRT-PCR
qPCR data differed for HCT-116 cells and colon CSCs, both by time and chemotherapeutic agents used. The optimum data were observed at 240 h of incubation. In colon CSCs, larger reductions in gene expression levels were observed at 240 h treatment when 5-FU was used as monotherapy. The use of oxaliplatin as a single agent affected gene expression to a lesser extent, while the combination was also sufficient but not as effective as 5-FU (Figure 5). In HCT-116 cells, the results were similar after incubation with 5-FU and 5-FU and oxaliplatin combined. Here, monotherapy with oxaliplatin was not satisfactory. Remarkable data were also obtained after 144 h of treatment of HCT-116 cells. During this time period, the separate administration of 5-FU and oxaliplatin led to a decrease in expression of up to 2-fold in the majority of genes, in comparison with their combination (Figure 6); the highest reduction was observed in thymidylate synthase (TYMS) following monotherapy with oxaliplatin for 144 h, while that in dihydrofolate reductase (DHFR) occurred following 240 h of treatment with either 5-FU alone or in combination with oxaliplatin. DNA (cytosine-5)-methyltransferase 1 (DNMT1) expression was decreased by 1.33-fold by 5-FU, 1.24-fold by oxaliplatin and 1.04-fold by their combination, while the greatest reduction in serine hydroxymethyltransferase (SHMT1) expression was observed after 240 h of treatment with the drug combination. However, DNA excision repair protein (ERCC1) expression was decreased at 192 h by the drug combination. Finally, dihydropyrimidine dehygrogenase (DPYD) expression was reduced by 1.91-fold at 144 h of incubation with 5-FU. DPYD was not expressed in colon CSCs. Regarding TYMS and DHFR, the optimum data were observed at 240 h for 5-FU (2.75-fold and 3.61-fold reduction, respectively). For DNMT1, SHMT1, and ERCC1, a greater reduction was noted after 192 h of incubation with oxaliplatin (4.17- fold, 4.08-fold and 3.67-fold, respectively). The administration of 5-FU after 240 h also had a remarkable effect but at lower levels. Nanog homeobox protein (NANOG) gene expression was decreased by 3.26-fold after 192 h
Figure 3. Viability assays of colon CSCs after 72 h, 144 h, 192 h and 240 h of incubation. Three different assays were performed to measure viability: MTT, SRB and CV assays. The values represent the percentage of dead cells.
of monotherapy with oxaliplatin in colon CSCs. Moreover, the single use of 5-FU led to a 2.35-fold decrease, while the combination treatment did not demonstrate significant results.
3.4. Cell Number
The number of viable cells was measured using a Nucleo Counter NC-100 image cytometer after administration of the drugs. The data were time-dependent for each cell line. After 72 h of incubation, the maximum number of dead cells was higher for the combination of oxaliplatin and 5-FU in both cell lines. 5-FU as monotherapy and FOLFOX gave much better results than oxaliplatin alone at 144 h treatment. The same results were observed in HCT-116 cells after 192 h of incubation. However, in colon CSCs, administration of 5-FU alone led to a significant decrease in viable cells. Oxaliplatin monotherapy after ten days dramatically reduced the number of viable cells in colon CSCs, while in HCT-116 cells, single administration either of 5-FU or oxaliplatin affected the cell population more significantly (Figure 7).
4. Discussion
There has been great progress in the development of CRC treatment; however, a high percentage of patients relapse after treatment, indicating that improved treatments are still required. Drug resistance is the biggest issue in increasing the efficacy of chemotherapy. Based on the CSC hypothesis, which suggests that rare populations of tumor-initiating cells, among others, may lead to resistance to therapy, the present study investigated the effect of 5-FU and oxaliplatin chemotherapeutic drugs administered in combination or as monotherapy in such cells.
Figure 4. Viability assays of HCT-116 cells after 72 h, 144 h, 192 h and 240 h of incubation. Three different assays were performed to measure viability: MTT, SRB and CV assays. The values represent the percentage of dead cells.
5-FU is a pyrimidine analog that inhibits thymidylate synthase. 5-FU causes cell cycle arrest in the post-G1/ pre-S phase and induces apoptosis by inhibiting DNA synthesis and by interacting directly or indirectly with several enzymes [26] . 5-FU enters the cell by a facilitated nucleobase transporter and is converted to its main active metabolites (FdUMP, FdUTP, and FUTP) by a complex metabolic pathway. It is thought to be cytotoxic to tumor cells via three potential mechanisms: inhibition of thymidylate synthase by FdUMP; incorporation of FdUTP into DNA; and incorporation of FUTP into RNA [27] . In the present study, genes encoding enzymes involved in these reactions included TYMS, SHMT1, DNMT1, DPYD and DHFR. TYMS catalyzes the methylation of deoxyuridylate to deoxythymidine, while DPYD is responsible for the degradation of uracil and thymine. SHMT1 plays an important role in nucleic acid biosynthesis, and DHFR reduces dihydrofolic acid to tetrahydrofolic acid. Tetrahydrofolate and its derivatives are essential for purine and thymidylate synthesis [28] - [31] . Previous studies in CRC demonstrated that the response to 5-FU varies in CRC cell lines [32] . In this study, it is remarkable that DPYD is not expressed in CSCs but only in HCT-116 cells. However, the gene expression disappaers after 192 or 240 h of incubation. This is also supported by a previous study, which demonstrated that colorectal tumors responding to 5-FU have low gene expression levels of dihydropyrimidine dehydrogenase [33] . The absence of DPYD in CRC stem cells could explain the effect of 5-FU; however it does not explain why monotherapy seems to be better than combination therapy. The effect of oxaliplatin was tested by studying the expression of ERCC1, a gene that encodes a DNA excision repair protein. According to a previous study, high expression of ERCC1 is correlated with resistance to platinum chemotherapy, as platinum compounds form crosslinks in DNA that inhibit DNA synthesis [34] . Previous studies have suggested FOLFOX in combination with other regimens such as curcumin to treat CRC cells [35] [36] .
The present work is an in vitro pharmacodynamic study that attempted to compare the effect of 5-FU and oxa- liplatin or their combination in HCT116 CRC cells and a colon CSC line. Data were generated by treating the
Figure 5. Gene expression analysis of colonl CSCs. The data were normalized according to control sample at 72 h. The data are presented in log2 ratio, thus values higher than 0 indicate increases in gene expression, and decreases if the value is lower than 0.
Figure 6. Gene expression analysis of HCT-116 cells. The data were normalized according to control sample at 72 h. The data are presented in log2 ratio, thus values higher than 0 indicate increases in gene expression, and decreases if the value is lower than 0.


Figure 7. Cell number pre- and post-treatment. The viability was tested with PI stain. (a) Colon CSCs; (b) HCT-116.
two different populations with the two drugs separately or in combination over a time window of 10 days (240 h). Despite the fact that these two cell population had totally different hallmarks, the results from the viability assays showed that the cell responses were almost comparable. According to the gene expression data, single use of 5-FU seemed to be more effective than in combination with oxaliplatin for both cell lines. However, there were differences in the response time: in CSCs, better results appeared after 192 h and 240 h treatment, while in HCT-116, 5-FU was more effective at 144 h. These observations may explain the resistance of CSCs over short incubation times, in contrary to HCT-116, where there was greater inhibition at shorter times. Specifically, in CSCs there was an increase in gene expression at shorter incubation times, which then decreased. Concerning each specific gene, no linearity was evident among them even though they are involved in similar pathways. However, in viability assays the percentage of dead cells increased in correlation with the incubation time.
In conclusion, the present study demonstrated that single use of 5-FU might have a better effect on CRC and colon CSCs compared with oxaliplatin or their combination. The viability of cells was decreased in both cell lines, while the gene expression pattern differed in CSCs. Moreover, gene expression profile should be studied for at least 10 days to ensure reliable results. In summary, this is the first report that monotherapy may be more effective; however, further larger studies are required.
Acknowledgements
None.
References
- Haggar, F.A. and Boushey, R.P. (2009) Colorectal Cancer Epidemiology: Incidence, Mortality, Survival, and Risk Factors. Clinics in Colon and Rectal Surgery, 22, 191-197. http://dx.doi.org/10.1055/s-0029-1242458
- Cunningham, D., Atkin, W., Lenz, H.-J., Lynch, H.T., Minsky, B., Nordlinger, B. and Starling, N. (2010) Colorectal Cancer. The Lancet, 375, 1030-1047. http://dx.doi.org/10.1016/S0140-6736(10)60353-4
- Wasserberg, N. and Kaufman, H.S. (2007) Palliation of Colorectal Cancer. Surgical Oncology, 16, 299-310. http://dx.doi.org/10.1016/j.suronc.2007.08.008
- Chau, I. and Cunningham, D. (2002) Adjuvant Therapy in Colon Cancer: Current Status and Future Directions. Cancer Treatment Reviews, 28, 223-236. http://dx.doi.org/10.1016/S0305-7372(02)00047-6
- de Gramont, A., Figer, A., Seymour, M., Homerin, M., Hmissi, A., Cassidy, J., Boni, C., Cortes-Funes, H., Cervantes, A., Freyer, G., et al. (2000) Leucovorin and Fluorouracil with or without Oxaliplatin as First-Line Treatment in Advanced Colorectal Cancer. Journal of Clinical Oncology, 18, 2938-2947.
- Shim, B.Y., Lee, K.M., Cho, H.-M., Kim, H.J., Cho, H.J., Yang, J., Kim, J.-G. and Kim, H.-K. (2005) Oxaliplatin/ 5-FU without Leucovorin Chemotherapy in Metastatic Colorectal Cancer. Cancer Research and Treatment, 37, 212- 215. http://dx.doi.org/10.4143/crt.2005.37.4.212
- Dean, M., Fojo, T. and Bates, S. (2005) Tumour Stem Cells and Drug Resistance. Nature Reviews Cancer, 5, 275-284. http://dx.doi.org/10.1038/nrc1590
- Woodward, W.A., Chen, M.S., Behbod, F., Alfaro, M.P., Buchholz, T.A. and Rosen, J.M. (2007) WNT/Beta-Catenin Mediates Radiation Resistance of Mouse Mammary Progenitor Cells. Proceedings of the National Academy of Sciences of the United States of America, 104, 618-623. http://dx.doi.org/10.1073/pnas.0606599104
- Liu, G.T., Yuan, X.P., Zeng, Z.H., Tunici, P., Ng, H., Abdulkadir, I.R., Lu, L.Z., Irvin, D., Black, K.L. and Yu, J.S. (2006) Analysis of Gene Expression and Chemoresistance of CD133+ Cancer Stem Cells in Glioblastoma. Molecular Cancer, 5, 67. http://dx.doi.org/10.1186/1476-4598-5-67
- Dean, M. (2009) ABC Transporters, Drug Resistance, and Cancer Stem Cells. Journal of Mammary Gland Biology and Neoplasia, 14, 3-9. http://dx.doi.org/10.1007/s10911-009-9109-9
- Liu, Y.B., Peterson, D.A., Kimura, H. and Schubert, D. (1997) Mechanism of Cellular 3-(4,5-Dimethylthia- zol-2-yl)-2,5- diphenyltetrazolium bromide (MTT) Reduction. Journal of Neurochemistry, 69, 581-593. http://dx.doi.org/10.1046/j.1471-4159.1997.69020581.x
- Stockert, J.C., Blazquez-Castro, A., Canete, M., Horobin, R.W. and Villanueva, A. (2012) MTT Assay for Cell Viability: Intracellular Localization of the Formazan Product Is in Lipid Droplets. Acta Histochemica, 114, 785-796. http://dx.doi.org/10.1016/j.acthis.2012.01.006
- van Meerloo, J., Kaspers, G.J. and Cloos, J. (2011) Cell Sensitivity Assays: The MTT Assay. Cancer Cell Culture Methods in Molecular Biology, 731, 237-245. http://dx.doi.org/10.1007/978-1-61779-080-5_20
- Sylvester, P.W. (2011) Optimization of the Tetrazolium Dye (MTT) Colorimetric Assay for Cellular Growth and Viability. Drug Design and Discovery Methods in Molecular Biology, 716, 157-168. http://dx.doi.org/10.1007/978-1-61779-012-6_9
- Liu, Y.B. and Nair, M.G. (2010) An Efficient and Economical MTT Assay for Determining the Antioxidant Activity of Plant Natural Product Extracts and Pure Compounds. Journal of Natural Products, 73, 1193-1195. http://dx.doi.org/10.1021/np1000945
- Ashraf, H., Taherian, A. and Kerdar, A.N. (2010) Evaluation of Cytotoxicity of Two Root Canal Filling Materials by MTT Assay. Australian Endodontic Journal, 36, 24-28. http://dx.doi.org/10.1111/j.1747-4477.2009.00173.x
- Han, F.X., Lin, H. and Ru, L. (2009) MTT Assay for Detecting 5-Fluorouracil Chemosensitivity of Human Breast Carcinoma Cell Line. Journal of Southern Medical University, 29, 97-99.
- Vichai, V. and Kirtikara, K. (2006) Sulforhodamine B Colorimetric Assay for Cytotoxicity Screening. Nature Protocols, 1, 1112-1116. http://dx.doi.org/10.1038/nprot.2006.179
- Voigt, W. (2005) Sulforhodamine B Assay and Chemosensitivity. Chemosensitivity Methods in Molecular Medicine, 110, 39-48. http://dx.doi.org/10.1385/1-59259-869-2:039
- Skehan, P., Storeng, R., Scudiero, D., Monks, A., McMahon, J., Vistica, D., Warren, J.T., Bokesch, H., Kenney, S. and Boyd, M.R. (1990) New Colorimetric Cytotoxicity Assay for anticancer-Drug Screening. Journal of the National Cancer Institute, 82, 1107-1112. http://dx.doi.org/10.1093/jnci/82.13.1107
- Houghton, P., Fang, R., Techatanawat, I., Steventon, G., Hylands, P.J. and Lee, C.C. (2007) The Sulphorhodamine (SRB) Assay and Other Approaches to Testing Plant Extracts and Derived Compounds for Activities Related to Reputed Anticancer Activity. Methods, 42, 377-387. http://dx.doi.org/10.1016/j.ymeth.2007.01.003
- Bonnekoh, B., Wevers, A., Jugert, F., Merk, H. and Mahrle, G. (1989) Colorimetric Growth Assay for Epidermal Cell Cultures by Their Crystal Violet Binding Capacity. Archives of Dermatological Research, 281, 487-490. http://dx.doi.org/10.1007/BF00510085
- Ishiyama, M., Tominaga, H., Shiga, M., Sasamoto, K., Ohkura, Y. and Ueno, K. (1996) A Combined Assay of Cell Viability and in Vitro Cytotoxicity with a Highly Water-Soluble Tetrazolium Salt, Neutral Red and Crystal Violet. Biological and Pharmaceutical Bulletin, 19, 1518-1520. http://dx.doi.org/10.1248/bpb.19.1518
- Grady, J.E., Lummis, W.L. and Smith, C.G. (1960) An Improved Tissue Culture Assay. III. Alternate Methods for Measuring Cell Growth. Cancer Research, 20, 1114-1117.
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25, 402-408. http://dx.doi.org/10.1006/meth.2001.1262
- Yoshikawa, R., Kusunoki, M., Yanagi, H., Noda, M., Furuyama, J.-I., Yamamura, T. and Hashimoto-Tamaoki, T. (2001) Dual Antitumor Effects of 5-Fluorouracil on the Cell Cycle in Colorectal Carcinoma Cells: A Novel Target Mechanism Concept for Pharmacokinetic Modulating Chemotherapy. Cancer Research, 61, 1029-1037.
- Longley, D.B., Harkin, D.P. and Johnston, P.G. (2003) 5-Fluorouracil: Mechanisms of Action and Clinical Strategies. Nature Reviews Cancer, 3, 330-338. http://dx.doi.org/10.1038/nrc1074
- Jensen, S.A., Vainer, B., Witton, C.J., Jørgensen, J.T. and Sørensen, J.B. (2008) Prognostic Significance of Numeric Aberrations of Genes for Thymidylate Synthase, Thymidine Phosphorylase and Dihydrofolate Reductase in Colorectal Cancer. Acta Oncologica, 47, 1054-1061. http://dx.doi.org/10.1080/02841860801942158
- Ichikawa, W., Uetake, H., Shirota, Y., Yamada, H., Nishi, N., Nihei, Z., Sugihara, K. and Hirayama, R. (2003) Combination of Dihydropyrimidine Dehydrogenase and Thymidylate Synthase Gene Expressions in Primary Tumors as Predictive Parameters for the Efficacy of Fluoropyrimidine-Based Chemotherapy for Metastatic Colorectal Cancer. Clinical Cancer Research, 9, 786-791.
- Steck, S.E., Keku, T., Butler, L.M., Galanko, J., Massa, B., Millikan, R.C. and Sandler, R.S. (2008) Polymorphisms in Methionine Synthase, Methionine Synthase Reductase and Serine Hydroxymethyltransferase, Folate and Alcohol Intake, and Colon Cancer Risk. Journal of Nutrigenetics and Nutrigenomics, 1, 196-204. http://dx.doi.org/10.1159/000136651
- DiPaolo, A. and Chu, E. (2004) The Role of Thymidylate Synthase as a Molecular Biomarker. Clinical Cancer Research, 10, 411-412. http://dx.doi.org/10.1158/1078-0432.CCR-1198-03
- Apostolou, P., Toloudi, M., Chatziioannou, M., and Papasotiriou, I. (2011) Predictive Value of Molecular-Cellular Methodologies for Response to Chemotherapy with CisPlatin/5Fu in Colon Tumours. Journal for Clinical Studies, 3, 28-32.
- Salonga, D., Danenberg, K.D., Johnson, M., Metzger, R., Groshen, S., Tsao-Wei, D.D., Lenz, H.J., Leichman, C.G., Leichman, L., Diasio, R.B., et al. (2000) Colorectal Tumors Responding to 5-Fluorouracil Have Low Gene Expression Levels of Dihydropyrimidine Dehydrogenase, Thymidylate Synthase, and Thymidine Phosphorylase. Clinical Cancer Research, 6, 1322-1327.
- Bellmunt, J., Paz-Ares, L., Cuello, M., Cecere, F.L., Albiol, S., Guillem, V., Gallardo, E., Carles, J., Mendez, P., de la Cruz, J.J., et al. (2007) Gene Expression of ERCC1 As a novel prognostic marker in Advanced Bladder Cancer Patients Receiving Cisplatin-Based Chemotherapy. Annals of Oncology, 18, 522-528. http://dx.doi.org/10.1093/annonc/mdl435
- Yu, Y.J., Kanwar, S.S., Patel, B.B., Nautiyal, J., Sarkar, F.H. and Majumdar, A.P. (2009) Elimination of Colon Cancer Stem-Like Cells by the Combination of Curcumin and FOLFOX. Translational Oncology, 2, 321-328. http://dx.doi.org/10.1593/tlo.09193
- Patel, B.B., Sengupta, R., Qazi, S., Vachhani, H., Yu, Y., Rishi, A.K. and Majumdar, A.P. (2008) Curcumin Enhances the Effects of 5-Fluorouracil and Oxaliplatin in Mediating Growth Inhibition of Colon Cancer Cells by Modulating EGFR and IGF-1R. International Journal of Cancer, 122, 267-273. http://dx.doi.org/10.1002/ijc.23097
NOTES
*Corresponding author.