Journal of Environmental Protection
Vol. 3 No. 12 (2012) , Article ID: 25690 , 21 pages DOI:10.4236/jep.2012.312181
Prokaryotic Horizontal Gene Transfer in Freshwater Lakes: Implications of Dynamic Biogeochemical Zonation
![]()
School of Geography and Earth Sciences, McMaster University, Hamilton, Canada.
Email: warrenl@mcmaster.ca
Received September 13th, 2012; revised October 17th, 2012; accepted November 12th, 2012
Keywords: Gene Transfer; Freshwater; Water Quality; Prokaryotic; Biogeochemistry
ABSTRACT
The highly adaptive nature of prokaryotic communities in the face of changing environmental conditions reflects in part their ability to share advantageous genetic information through horizontal gene transfer (HGT). Natural freshwater lacustrine (lake) systems are a vital and finite resource, and the influence of HGT on their quality (e.g. enabling the spread of antibiotic resistance and xenobiotic catabolism genes) is likely significant. Laboratory and in situ studies indicate that the dynamic physical, chemical, and biological conditions that structure freshwater systems can influence HGT within freshwater prokaryotic communities. Thus, understanding how biogeochemical parameters impact HGT in freshwater lakes is an emerging knowledge gap with potential implications for ecosystem and human health on a global scale. In this review, we provide a general synopsis of what is known about HGT in freshwater prokaryotic communities, followed by an integrated summary of current knowledge identifying how biogeochemical factors may influence prokaryotic HGT in freshwater lacustrine systems.
1. Introduction
Intercellular horizontal gene transfer (HGT), the movement of genetic information between cells by mechanisms operating independently of cell division (i.e. vertical gene transfer), has contributed to the macroevolution of prokaryotes and their attainment of tremendous structural and functional diversity [1,2]. Further, the highly adaptive nature of prokaryotic communities in the face of changing environmental conditions reflects in part their ability to share advantageous genetic information through HGT. The horizontal spread of antibiotic resistance genes among pathogenic bacteria [3], catabolic genes among bacteria inhabiting soil contaminated with organic pollutants [4,5], and foreign genes carried by genetically engineered organisms into natural prokaryotic communities [6,7] highlights the breadth of the environmental impact of prokaryotic HGT.
Prokaryote-mediated biogeochemical processes are fundamental to aquatic ecosystem functioning [8], such that functional community changes resulting from HGT may have substantial repercussions. The dissemination of introduced genetic information in aquatic prokaryotic communities via HGT can potentially impact water quality by such diverse processes as altering their health hazard potential (e.g. acquisition of introduced pathogenicity-enhancing genes) or ability to carry out particular metabolic pathways (e.g. degradation of recalcitrant organic pollutants). Natural freshwater lacustrine (lake) systems are a vital and finite resource, collectively accounting for 94% of the world’s fresh surface water by volume [9]. As water use increases and water quality decreases, it is increasingly clear that our freshwater resources have gone beyond a critical status on a global scale, where many countries are without adequate safe and secure water supplies. Reflecting the links between water quality and HGT, the growing spread of clinically derived antibiotic resistance genes, an emerging class of environmental contaminants [10,11], in freshwater prokaryotic communities by HGT has been the focus of recent attention [12-14]. Laboratory and in situ studies indicate that the dynamic physical, chemical, and biological conditions that structure freshwater systems can influence the nature, extent and rates of HGT within freshwater prokaryotic communities. However, mechanistic understanding of how biogeochemical parameters impact HGT in freshwater lakes is an emerging knowledge gap for both ecosystem and human health on a global scale. This review provides an integrated summary of current knowledge identifying how biogeochemical factors influence the horizontal flow of genetic information in freshwater lacustrine prokaryotic communities.
2. Prokaryotic Horizontal Gene Transfer in Freshwater Systems
Intercellular HGT consists of the acquisition, stable assimilation, and expression of foreign DNA via specific cellular mechanisms. Foreign DNA derived from a donor cell is introduced into a recipient cell predominantly via 1) direct intercellular exchange (conjugation); 2) virusmediated transport (transduction); or 3) uptake from the extracellular environment (transformation). As the emphasis of this review is on the regulation of HGT by variable biogeochemical parameters within freshwater lacustrine systems, the reader is referred to specific reviews covering conjugation [15], transformation [16], and transduction [17] for more detailed discussion of the mechanisms themselves.
Single-cell DNA sequencing of bacteria inhabiting freshwater lakes has revealed evidence for inter-phyla HGT [18] and the three principal mechanisms of intercellular HGT have been observed in situ in freshwater environments (e.g. [19-21]); however little is known about the magnitude and relative contributions of each mechanism, the indigenous participant prokaryotes, or the overall impact of freshwater HGT on water quality and prokaryotic microevolution. In the Baltic Sea, genomic sequence analysis of Shewanella baltica isolates from various depths within the water column indicated that genes enabling adaptation to particular environmental conditions (e.g. anaerobic metabolism genes) were horizontally exchanged to a greater extent at more similar depths (i.e. biogeochemical conditions) as well as over seasonal time frames [22]. This finding suggests that HGT rates may vary with water column depth-dependent biogeochemical zonation within freshwater zystems, and that the horizontal transfer of ecologically relevant genes occurs over timescales that are sufficiently rapid to have appreciable system impacts.
Most studies of aquatic HGT have been carried out in microcosms, which while capable of providing approximate in situ transfer rates [23] and permitting some control of individual environmental parameters, cannot entirely replicate the complexity of in situ conditions [24]. These studies have predominantly utilized culture-dependent approaches, although more recent investigations have utilized molecular microscopy approaches to directly assay HGT within freshwater prokaryotic communities, albeit under laboratory conditions. Shintani et al. [25] detected the conjugative transfer of a plasmid tagged with a fluorescent protein gene in river water samples. Kenzaka et al. [26] directly detected transduction in a freshwater river prokaryotic community mediated by three Escherichia coli-infecting phages, including one previously isolated from a freshwater river, using a DNA amplification-based in situ microscopy technique. A similar approach was used to detect the uptake and expression of an introduced plasmid tagged with a fluorescent protein gene by indigenous prokaryotic communities in two Japanese rivers up to three weeks following plasmid introduction [27,28]. A small number of field studies of freshwater HGT have been performed to date, and will be briefly described in the following section.
2.1. Conjugation
Conjugative plasmids have been isolated from bacterial communities in freshwater lakes [29] and rivers [30-33], indicating that conjugation is common in freshwater systems. O’Morchoe et al. [20] detected conjugative plasmid transfer in situ between strains of the freshwater bacterium Pseudomonas aeruginosa introduced into a semi-permeable chamber suspended in the upper pelagic waters of a freshwater lake. Conjugative plasmid transfer has also been demonstrated in situ in biofilms on the submerged surfaces of river stones (epilithon) between introduced strains of P. aeruginosa [30-32] and from an indigenous freshwater epilithic prokaryotic community to an introduced recipient Pseudomonas strain [32,34].
2.2. Transduction
Viruses are ubiquitous in freshwater environments, typically exceeding bacteria in concentration by an order of magnitude [35]. Phages (for the purposes of this review, viruses that infect prokaryotes) constitute the majority of freshwater viruses, exhibit high diversity, and consistently infect a substantial portion of freshwater prokaryotic cells, indicating that transduction is an important form of HGT in freshwater systems [36]; however to date this remains largely unconstrained. Most known species of bacteria can be infected by phages, some of which can infect more than one species or even genus [37]. Many prokaryotic cells in freshwater systems are lysogenic, meaning that phage DNA capable of being reactivated to produce viral particles has been integrated into their chromosomes as inducible prophage [19]. Induction of prophage in lysogenic cells can potentially stimulate both transduction and transformation by increasing the availability of phage particles and extracellular DNA, respectively, which are released following cell lysis. Phage particles may be rapidly inactivated in aquatic environments [36], so the frequency and induction of lysogeny in prokaryotic cells are important determinants of transduction potential in aquatic environments [19].
Transduction of chromosomal and plasmid DNA has been demonstrated in situ between strains of Pseudomonas aeruginosa intentionally infected with species-specific phages and introduced into a semi-permeable chamber suspended in the upper pelagic waters of a freshwater lake [19,38-40]. Transduction between introduced lysogenic bacterial strains following natural induction of lysogeny has also been demonstrated in situ in river epilithic biofilms [41].
2.3. Transformation
Transformation in aquatic environments is contingent upon the availability of genetically competent (able to uptake extracellular DNA) prokaryotes and extracellular DNA sharing homology with the intracellular DNA of recipient competent cells. The reader is referred to Nielsen et al. [42] for a detailed review of the release and fate of extracellular DNA in aquatic environments. Dissolved (<0.2 µm) extracellular DNA including fragments of sufficient size to encode prokaryotic genes has been measured at concentrations on the order of 10-1 to 101 µg/l in bulk water from freshwater lakes, reservoirs, ponds, and rivers [43-45], with transformable free DNA molecules (as opposed to viral or colloid-associated DNA) constituting a major form of dissolved DNA in aquatic systems [44,46]. Extracellular DNA is also a component of the extracellular polymeric substances that surround prokaryotic cells in biofilms [47]. Sources of prokaryotic extracellular DNA in aquatic systems include excretion by living cells and release from dead cells following grazing or lysis (e.g. phage-induced) [48,49]. Aquatic extracellular DNA is degraded by extracellular and cell-associated nucleases, genotoxic chemicals, as well as solar ultraviolet radiation [50]. In particular, the activity of extracellular nucleases can strongly influence the persistence of extracellular DNA in freshwater environments [51].
In situ transformation in freshwater systems remains poorly characterized. The transformation of an introduced recipient strain of Acinetobacter calcoaceticus by co-introduced extracellular DNA from cell lysates or intact donor A. calcoaceticus cells has been demonstrated in situ in river epilithic biofilms [21].
While these studies indicate the occurrence of prokaryotic HGT within freshwater systems, their relevance is limited by two important factors. Firstly, they were restricted to the lighted and well-oxygenated temperate environments of either biofilms on the submerged surfaces of river stones or the upper offshore open waters of lakes, poorly representing the range of habitats and extensive biogeochemical variation found within freshwater systems. Secondly, they employed laboratory-grown strains of freshwater-inhabiting species of Acinetobacter or Pseudomonas bacteria (both belonging to the Pseudomonadales order of Gammaproteobacteria) as donors (cells, DNA, or phage particles from lysogens) and/or recipients. In stark contrast, environmental prokaryotic communities are well known to be highly diverse and remain largely uncharacterized [52,53] and Gammaproteobacteria are typically not major constituents of freshwater lake bacterial communities [54]. Of key significance, prokaryotes utilize a wide variety of metabolic pathways to obtain energy and organic carbon for growth. Autotrophic prokaryotes (e.g. photoautotrophs, sulfuroxidizing lithoautotrophs) are widespread in freshwater environments, and heterotrophs capable of using terminal electron acceptors other than oxygen (e.g. ironand sulphur-reducing bacteria) can dominate prokaryotic communities in oxygen-poor and anoxic regions (e.g. bottom sediments and bottom waters of productive lakes). These metabolic guilds are capable of substantially altering their local environments (i.e. generating biogeochemical zonation) such that they and their neighbours encounter distinct biogeochemical conditions [55-58] that may differentially influence HGT within freshwater systems. Further, these prokaryotes are often challenging to grow under laboratory conditions owing to their oxygen, nutrient and/or consortial requirements and so are likely underrepresented among HGT studies to date. The potential influence of the particular metabolism a prokaryote is carrying out on HGT remains essentially undefined. Thus, the characterization of HGT based principally on only two species of bacteria of the same order in a highly limited range of habitats and biogeochemical conditions means that the full context of freshwater HGT has yet to be determined.
3. Biogeochemical Factors Influencing HGT in Freshwater Lakes
The biogeochemical conditions found in freshwater lacustrine systems can potentially influence all mechanisms of HGT (Figure 1(a)). Many of the factors known to influence HGT typically exhibit spatial and temporal variation among and within freshwater lakes. While the impacts of most of these factors have been individually assessed with respect to HGT, most studies have assessed impacts in isolation, impeding understanding of the role of environmental interactions or spatial/temporal variability of these factors within freshwater lakes.
Field studies of freshwater HGT to date have been highly limited in their scope of system characterization, and with the exception of a small number of studies examining temperature [21,30-32] or suspended particulate matter concentrations [40], lack data linking biogeochemical parameters to observed HGT rates in the particular freshwater compartment investigated. Most studies of biogeochemical factors influencing HGT have been conducted in vitro, utilizing a limited number of easily culturable chemoorganoheterotrophic bacteria grown aerobically in nutrient rich media under highly constrained laboratory conditions. Such an approach is poorly representative of the myriad permutations of biogeochemical conditions to which freshwater prokaryotic communities are typically exposed, as well as their substantial phylogenetic and metabolic diversity. This has limited the generation of new insight into environmenttally linked HGT and reduced the relevance of any findings for environmental systems. Here we provide a summary of what has been shown for these factors in terms of their influence on HGT to date, providing context for what is known about their variability within freshwater lacustrine systems where possible.
3.1. Temperature
Conjugative plasmid transfer rates for laboratory cultures of Gram-negative bacteria have been observed to be temperature sensitive, potentially reflecting the limited expression of conjugative machinery within certain temperature ranges independent of optimal temperature ranges for growth [59,60]. The relationship between temperature, optimal growth, and HGT in natural freshwater prokaryotic communities is likely to be appreciably more complex. In stream epilithic biofilms, rates of conjugation and transformation between laboratory strains of bacteria have been positively correlated with temperature over seasonal timeframes, suggesting that HGT increases during summer months in temperate climates, particularly in surface waters warmed by solar radiation, and consistently occurs with greater frequency in warmer climates [21,30-32]. However, the observed influence of temperature on HGT in these studies may simply reflect the optimal temperature range for growth of the mesophilic bacterial strains that were used. Temperature was positively correlated with the abundance, production, and specific growth rate of marine heterotrophic bacteria throughout Chesapeake Bay during non-summer seasons [61], suggesting that it may similarly control the growth of prokaryotes in temperate freshwater lakes and their metabolic capacity for participating in HGT.
Transduction may be influenced by temperature, but the direction and magnitude of this relationship are currently poorly defined. Viral decay rates are generally reduced at lower temperatures in aquatic systems [36], suggesting that transduction rates are greater in colder water due to greater phage availability. However, viral abundance has been significantly positively correlated with temperature in a thermally stratified freshwater reservoir [62] and prophage induction was demonstrated in vitro to increase by several orders of magnitude with an increase in temperature from 22˚C to 28˚C in the presence of ultraviolet radiation [63], so the relationship between temperature and transduction in freshwater systems remains unclear.
Transformation rates may be greater in cooler waters as the reduced activity of nucleases at lower temperatures increases the availability of transformable extracellular DNA [64]. Using water collected from different regions within a thermally stratified freshwater lake during summer, Matsui et al. [65] found that degradation of introduced exogenous plasmid DNA with a concurrent decrease in transformation efficiency was observed over several days in epilimnetic water (18.0˚C) but not in hypolimnetic water (7.8˚C), both held at the same temperature as measured in the field. Taken together, these observations indicate that rates of transformation may be higher in summer hypolimnetic regions of temperate lakes. However, what is not known is whether these findings are directly translatable to rates of transformation within the naturally occurring prokaryotic communities within these regions.
3.2. Dissolved Oxygen
As depth-related variation in the concentration of dissolved oxygen can often mirror thermal stratification in freshwater lakes (Figure 1(b)), it is inherently difficult to tease out the separate effects of temperature and oxygen concentration on freshwater HGT. Further, no in situ studies examining the influence of dissolved oxygen concentration on HGT in freshwater systems have been published to date. As discussed previously, under low oxygen and anoxic conditions prokaryotes often utilize terminal electron acceptors other than oxygen, and so may distinctly change their local environments, in turn influencing HGT. In a laboratory setting, the efficiency of competence development and frequency of transformation were both reduced in the facultative anaerobe Streptococcus pneumoniae under anaerobic conditions [66]. Similarly, lower rates of conjugation have been observed under anaerobic conditions relative to aerobic conditions in laboratory studies utilizing the facultative anaerobe Escherichia coli [67,68]. Conjugation rates were observed to increase 10-fold following increased aeration of a laboratory culture of E. coli [69]. Notably, these organisms are not typically dominant or key players in freshwater prokaryotic communities observed within freshwater systems and thus the relevance of these findings to environmental prokaryotes, many of which are autotrophic and/or obligate anaerobes, is unknown. However, collectively, these findings indicate that conjugation and transformation may be inhibited in anoxic zones, both in bulk (e.g. hypolimnion of highly productive thermally stratified lakes) and microenvironments (e.g. within biofilms), among facultatively anaerobic members of inhabitant prokaryote communities. Laboratory research clearly indicates that HGT among bacteria capable of both aerobic and anaerobic growth is influenced by dissolved oxygen concentration; however in situ studies are required to characterize this regulation in freshwater lacustrine systems.
3.3. Chemical Agents
A suite of natural and anthropogenic chemicals can
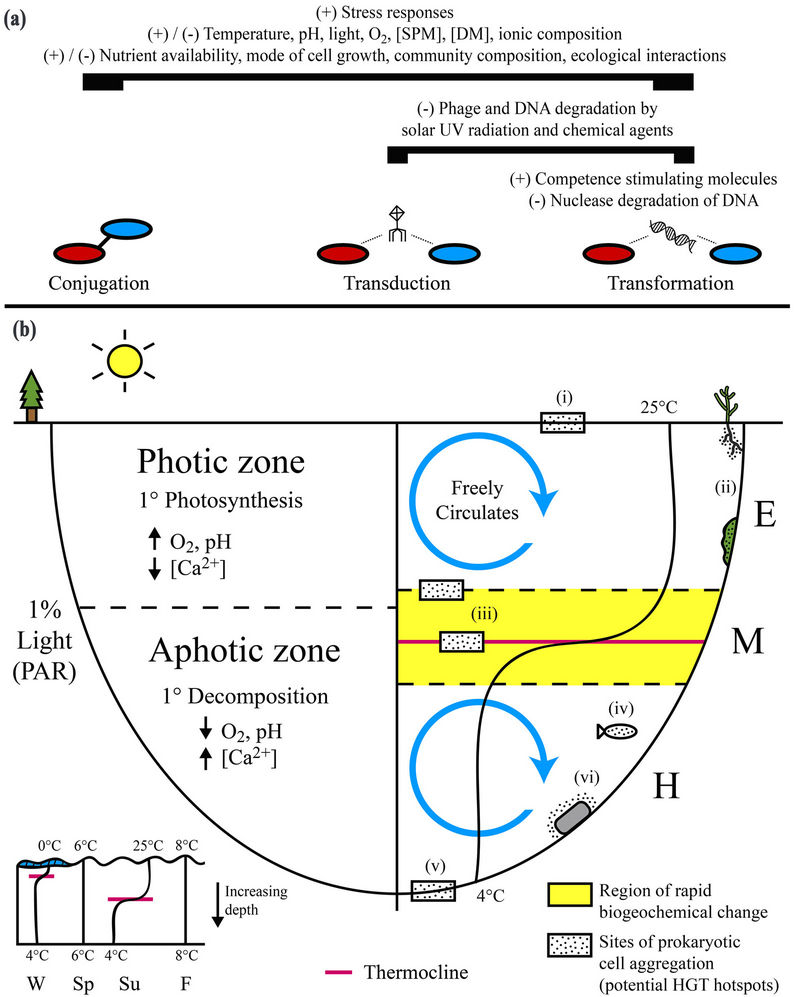
Figures 1. Physical, chemical, and biological parameters capable of regulating horizontal gene transfer (HGT) can vary appreciably over space and time within freshwater lacustrine systems. (a) Biogeochemical parameters as a whole remain poorly characterized with respect to their in situ influence on HGT. Many parameters may alternatively stimulate (+) or inhibit (−) all three principal mechanisms of gene transfer (conjugation, transduction, and transformation) in freshwater systems based on their magnitude and interactions with other parameters. In general, activation of stress responses can stimulate HGT. Degradation of phage particles and extracellular DNA, the external vehicles for transduction and transformation, respectively, can inhibit these mechanisms. Competence stimulating molecules, produced by prokaryotes and algae, can promote transformation; (b) Biogeochemical parameters capable of influencing horizontal gene transfer can vary appreciably within freshwater lakes. In particular, the vertical physical separation of chemical and biological processes by depth-dependent changes in light availability and thermal density can lead to the formation of distinct local environmental conditions that likely have a specific cumulative influence on HGT. In general, HGT is enhanced in surficial aggregations of metabolically active cells (i.e. biofilms). Sites representing potential hotspots for HGT within freshwater lakes include 1) the air-water interface; 2) aquatic plants and algae; 3) vertical zones of rapid density and subsequent biogeochemical change within the metalimnion (e.g. thermocline); 4) aquatic animals; 5) bottom sediments and the sediment-water interface; and 6) submerged abiotic surfaces (e.g. rocks). SPM = suspended particulate matter, DM = dissolved matter; PAR = photosynthetically active radiation; W = winter, Sp = spring, Su = summer, F = fall; E = epilimnion, M = metalimnion, H = hypolimnion.
potentially influence HGT in freshwater lakes, although their in situ effects largely remain uncharacterized. Chemicals can damage prokaryotic cells, phage particles, and extracellular DNA, limiting their participation in HGT. Detergents, mutagens, and antibiotics have been observed to inhibit conjugation in vitro, either directly or indirectly via induction of plasmid degradation and/or selective toxicity targeting plasmid-containing cells [70, 71]. Conversely, in vitro exposure to chemicals has been demonstrated to stimulate HGT between bacterial species otherwise unlikely to exchange genetic information, likely by inactivating restriction enzymes that normally limit interspecific HGT [71-73].
Genotoxic chemicals including antibiotics and trace elements can promote all three principal mechanisms of HGT in vitro by activating bacterial stress responses (e.g. SOS response), leading to the induction of competence [74,75], prophage [76,77], and expression of conjugation machinery [78]. In particular, prophage induction via activation of the SOS response in lysogenic strains of both Gram-positive and Gram-negative bacteria has been demonstrated in vitro for a variety of chemicals at sublethal concentrations including antibiotics, antineoplastic drugs, halogenated organics, polycyclic aromatic hydrocarbons, and trace elements [79-83]. SOS response activation was observed in cultures of the aquatic bacterium Vibrio cholerae following exposure to subinhibitory concentrations of most major groups of clinically used antibiotics [84], indicating that antibiotic-mediated stimulation of transduction can occur in freshwater prokaryotic communities. Collectively, in vitro investigations of the influence of chemical agents on HGT via bacterial stress response activation suggest that freshwater lake compartments containing biologically accessible genotoxic chemicals at concentrations sufficient to induce stress responses are sites of higher HGT rates. Supporting this conclusion, a genotoxicity assay based on prophage induction in Escherichia coli produced positive results for bulk water samples collected at sites along a freshwater river most highly contaminated by trace elements [85].
Horizontally mobile genetic elements including integrons, transposons, and plasmids were found to be more abundant in freshwater river sediments exposed to waste water containing relatively high concentrations of antibiotics [86]. Similarly, exposure to trace elements has been associated with increased abundance of IncP-1 plasmids in freshwater river sediments [87] and class 1 integrons in freshwater river epiphytic biofilms, bulk surface water, and bottom sediments [88,89]. The general extent of freshwater system contamination with chemicals has been positively correlated to the incidence of potentially mobile plasmids in bacterial isolates [90]. As the abundance of mobile genetic elements can be used as an indicator of relative HGT potential in prokaryotic communities [88], these findings support the notion that freshwater lake compartments with relatively high concentrations of antibiotics and trace elements may be hotspots for HGT.
Chemical agents capable of influencing HGT are principally introduced into freshwater lacustrine systems as a consequence of anthropogenic activities [91,92]. Generally speaking, the lowest concentrations of trace elements and antibiotics necessary to stimulate HGT in vitro are several orders of magnitude greater than concentrations typically found in freshwater environments [93,94], so it is unclear whether bacterial stress response activation by these chemicals is relevant to HGT regulation in most freshwater systems. However, chemicals may be sufficiently enriched in compartments within lakes that they can locally influence HGT. Trace elements and organic pollutants can accumulate within bottom sediments and at the sediment-water and air-water interfaces of aquatic systems [95-97], and aquatic biofilms can sorb a wide variety of inorganic and organic chemicals [47,98,99], suggesting that these compartments could be hotspots for HGT within freshwater lakes. Antibiotics are naturally produced by common freshwater microorganisms includeing Actinobacteria and filamentous fungi, and so may influence HGT in freshwater habitats in which these microorganisms are active and abundant. As discussed previously, prokaryotes are capable of a wide variety of metabolisms, and particular metabolisms may distinctly alter biogeochemical conditions, in turn influencing HGT, although this remains essentially undefined. For example, iron-reducing bacteria in anoxic lake bottom sediments can degrade recalcitrant organic pollutants and liberate trace elements through the dissolution of iron minerals [58], potentially resulting in their differential exposure to chemicals that can regulate HGT via activation of stress responses. Prokaryotes can also transform the redox state of trace elements such as arsenic, altering their mobility, bioavailability, and toxicity [100], and ultimately their influence on HGT.
3.4. Solar Ultraviolet Radiation
As with chemicals, solar ultraviolet radiation can potentially stimulate or inhibit all three principal mechanisms of HGT by activating bacterial stress responses [101] and damaging participating bacterial cells [102], phage particles [102-104], and DNA [105], respectively. In situ effects of solar ultraviolet radiation on HGT in freshwater lacustrine systems are not well characterized. Ultraviolet wavelengths are rapidly attenuated within water bodies and thus their impacts will largely be restricted to the upper 1 - 2 meters of a lake. Prophage induction in lysogenic prokaryotic cells following exposure to solar radiation has been observed in water samples collected from freshwater lakes [106,107]. Exposure of microcosms prepared in situ from seawater to solar radiation increased prokaryotic mortality and production of viral particles, suggesting an increase in transduction rates following prophage induction [37]. Consistent with the surficial limitation of ultraviolet radiation impacts, Tapper and Hicks [106] determined that the surface microlayer at the air-water interface of a freshwater lake was enriched in virus particles relative to the underlying epilimnion water, indicating the occurrence of appreciable prophage induction by the high intensity of solar ultraviolet radiation incident on the water surface. Ozonedriven increases in solar ultraviolet-B radiation over the last several decades with concurrent negative impacts on aquatic ecosystems [108] indicate that solar ultraviolet radiation will continue to increase in importance as a regulator of HGT in lacustrine systems.
3.5. Suspended Particulate Matter and Dissolved Matter
Suspended particulate matter and dissolved matter (typically operationally defined as materials suspended in the water column with diameters greater and less than 0.45 µm, respectively) can influence rates of HGT in freshwater lakes through a number of processes. Both contribute to the attenuation of solar radiation and thereby limit the vertical region in which it can directly influence HGT. In particular, dissolved organic matter, a major form of dissolved matter in most freshwater systems, can strongly reduce ultraviolet penetration [109]. Photo-oxidation mediated by solar ultraviolet-B radiation in humic (organic carbon rich) lakes was observed to be restricted to the top couple of centimeters [110], suggesting that the vertical region in which solar ultraviolet radiation can appreciably influence HGT is further restricted in systems with higher dissolved organic matter concentrations. Concentrations of dissolved organic matter are generally higher in highly biologically productive systems, reflecting their in situ production. Dissolved organic matter is also higher in lake systems impacted by anthropogenic organic-rich wastewater inputs (e.g. sewage, agricultural runoff) or those receiving significant wetland inputs. The influence of dissolved organic matter on HGT may be increasingly pronounced in Boreal Forest lake systems impacted by climate change, as melting permafrost is driving significant exports of organic matter via surrounding organic-rich wetlands to downstream lakes.
Conversely, the presence of suspended particulate matter can potentially stimulate all three principal mechanisms of HGT by providing surfaces for the adsorption of prokaryotic cells, phage particles and extracellular DNA, which stabilizes and protects these HGT participants from damage by extracellular enzymes, toxic chemicals, and solar ultraviolet radiation while also increasing the probability of cell-cell, DNA-cell, and phage-cell encounters and resultant HGT [40,51,111-114]. Furthermore, particulate-associated prokaryotes are typically more metabolically active than planktonic (free-living) prokaryotes [115,116], suggesting that the former can participate to a greater extent in HGT. Ripp and Miller [40] measured a 100-fold increase in the number of transductants when sterilized freshwater microcosms were prepared using water collected near a lake bottom and enriched in suspended particulate matter compared to surface water with little particulate matter. The addition of clay mineral particles has also been shown to enhance in vitro transduction rates [40]. The affinity of a phage particle for adsorption to a surface is dependent on the type of phage and surface involved [117]. Kernegger et al. [118] found that the abundances of adsorbed bacteria and viruses, as well as the ratio of adsorbed virus to adsorbed bacteria, were significantly higher for suspended organic particles compared to suspended mineral (inorganic) particles, suggesting that organic particles may be sites of higher HGT rates. Extracellular DNA adsorbs to clay minerals, sand particles, and natural organic matter [119] and in this form is available for transformation [111, 112,120,121]. The high affinity of DNA for organic and mineral phrases present in bulk in aquatic environments and the protection that adsorption confers against degradation by nucleases and toxic chemicals [119] inndicate that adsorbed DNA represents an important reservoir of transformable DNA in freshwater systems. Transformation frequencies of adsorbed DNA have been observed to be alternatively higher [111], lower [112], or similar [122] relative to dissolved DNA under laboratory conditions, and the type of DNA sorbent can influence transformation efficiency [122,123], so the net effect of DNA adsorption on transformation in aquatic systems is likely complex. Variability in the amount of different mineral and organic phases available for adsorption of cells, phages, and DNA within freshwater lacustrine systems may result in system and system compartment specific control of transformation rates (e.g. nutrient poor subglacial lakes with high suspended clay mineral concentrations compared to nutrient rich lakes with high concentrations of organic matter). Finer (smaller grain size) suspended matter possesses relatively greater surface area to volume ratios and typically stay in suspendsion longer, and so may enhance contact between HGT participants to a greater extent than more coarse particles due to a greater collective capacity for HGT participant adsorption. Plasmid transfer rates were observed to be greater in soils with high clay content (i.e. finer textured soils) [124], suggesting that HGT rates may be similarly enhanced in freshwater lakes with finer suspended matter (e.g. lakes receiving extremely fine glacial powder from runoff). Concurrent sorption of HGT participants and chemical agents capable of influencing gene transfer may further modulate the effect of suspended particulate matter on freshwater HGT. In freshwater lakes, suspended floc, which are aggregates of cells, organic matter and minerals, may be highly enriched in trace elements relative to bottom sediments and the bulk water phase [125, 126], potentially resulting in localized stress-driven HGT changes. At the lower end of the particle size scale, nanomaterials in water are capable of influencing HGT. Nanoalumina has been demonstrated to increase in vitro conjugative plasmid transfer across bacterial genera via increased stress and alteration of gene expression [127].
Concentrations of suspended particulate matter may vary substantially over time and space within freshwater lakes. Climatic variability (e.g. storms), anthropogenic point and non-point sources (e.g. combined sewer outfalls, agricultural runoff), phytoplankton blooms, as well as seasonal events including the turnover of stratified systems and springtime melting of snow and ice in temperate climates, may temporarily and locally or generally stimulate HGT by increasing concentrations of suspended particulate matter in freshwater systems via the resuspension of settled particles or increased particulate matter loads in inflowing waters. Concentrations of suspended particulate matter, and rates of HGT, may be relatively high in the metalimnion of thermally stratified lakes due to the accumulation of particulates from the epilimnion at the thermocline where maximal thermally related density differences inhibit mixing of water layers. Microbial communities are commonly concentrated in this region, taking advantage of the local build up of organic carbon and nutrients as well as the unique biogeochemical and energetic conditions that are often present. Similarly, suspended particulate matter is typically enriched at the air-water [128] and sediment-water interfaces of freshwater systems, and may also be generally elevated in the hypolimnion of stratified freshwater lakes due to internal seiches or large water circulation movements driving the resuspension of settled particles [129].
3.6. pH
No in situ studies examining the influence of pH on HGT in freshwater systems have been published to date. Rates of in vitro transformation and conjugation involving bacteria from freshwater river water or epilithic biofilms as donor cells were not significantly affected by pH in the range (6 - 8) found in most freshwater lakes [130-133]. In situ bacterial production was similar across acidified and limed humic lakes (pH range 5 - 7), suggesting that pH does not appreciably influence HGT via a general effect on prokaryotic metabolic activity [134]. However, freshwater pH may influence HGT by controlling the net surface charge present on prokaryotic cells, phage particles, and extracellular DNA, as well as the mineral and organic surfaces they adsorb to, altering surface-facilitated interactions between mediators of HGT and recipient cells and protecting HGT participants from degradation. Romanowski et al. [51] observed that the adsorption of plasmid DNA to sand and groundwater aquifer material in microcosms was inversely dependent on the pH of the solution (pH range 5 - 9), reflecting the increasing negative charge on mineral particle surfaces typically observed with increasing pH, which repulses the negatively charged DNA molecules [51]. This finding suggests that less DNA may be available for transformation in freshwater lacustrine systems and system compartments with higher pH due to the lack of protection from degradation conferred by adsorption to mineral surfaces. The bioavailability of trace elements, which dictates their potential for influencing HGT rates in freshwater prokaryotic communities, is also pH-dependent. The pH of a freshwater lake reflects both its chemical and biological characteristics, and is variable over depth dependent spatial scales as well as diel, seasonal and annual timeframes. This variability may be reflected in changing freshwater HGT rates. For example, diel variation in HGT may occur in lacustrine zones of high photosynthetic activity, as fluctuations in pH may be substantial (i.e. 1 - 2 pH unit shifts) over this timescale, with pH increasing occurring during lighted hours and decreasing at night when respiration dominates [135].
3.7. Ionic Composition
As with pH, the total dissolved ion concentration (salinity) of a freshwater environment can affect the adsorption of prokaryotic cells, phage particles, and extracellular DNA to mineral and organic surfaces, and in turn, the potential for HGT [36,51,136,137]. Freshwater systems, in contrast to marine systems, are highly dilute with the highest concentrations of the most abundant typical ion, Ca2+, in the mM range (in contrast to water itself at 25 M). The adsorption of DNA to sand and groundwater aquifer material in microcosms was enhanced in the presence of divalent cations, with concentrations for half-maximum DNA adsorption of 0.5 mM for Mg2+ and 0.4 mM for Ca2+ [51,112,120]. In vitro transformation of the soil bacterium Azotobacter vinelandii by DNA adsorbed to natural organic matter significantly increased in the presence of 1 mM Ca2+ [122]. Similarly, in vitro transformation of Bacillus subtilis was optimal with media containing 20 mM Mg2+ [138]. Together, these findings suggest that transformation rates in freshwater prokaryotic communities may be influenced by the concentrations of divalent cations in their local environment. Metal cations, particularly Ca2+ and Mg2+, are involved in competence development, and Ca2+ is necessary for transformation to occur in bacteria [139-141]. Trombe et al. [142] determined that 1 mM Ca2+ was sufficient to induce competence in vitro using Streptococcus pneumoniae, while Page and von Tigerstrom [139] found that 0.5 to 1 mM Ca2+ resulted in optimal competence induction in vitro using Azotobacter vinelandii. Transformation of Escherichia coli with plasmid DNA was demonstrated in vitro using fresh river water containing concentrations of Ca2+ as low as 1.4 mM, with rates of transformation generally being directly related to Ca2+ concentration [140]. The authors noted that rivers sampled in a calcareous area had Ca2+ concentrations of 2 - 2.5 mM, while those from a granitic area (minerals with low [Ca2+]) had concentrations of 0.3 - 1.5 mM, suggesting that transformation rates may be higher in lacustrine systems with drainage basins rich in calcium carbonate rock [140]. Concentrations of Ca2+ can also vary appreciably with depth in stratified hardwater (i.e. Ca2+ rich) lakes, with photosynthetically driven calcium carbonate (CaCO3) precipitation (associated with increasing pH) decreasing concentrations in the euphotic regions of the epilimnion and metalimnion during lighted hours, and decomposition (commonly decreases pH through CO2 generation which increases CaCO3 solubility) increasing concentrations in the hypolimnion. In highly productive hardwater lakes, Ca2+ concentrations can substantially decrease during periods of intense biological activity, which can strip out the Ca2+ in CaCO3 precipitates, potentially inhibiting transformation.
3.8. Prokaryotic Cell Density and Mode of Growth
Higher rates of HGT are typically found in more dense populations of prokaryotic cells, primarily due to the increased probability of contact between mediators of HGT and recipient cells [143]. Using a continuous culture model under conditions relevant to those found in freshwater environments, Replicon et al. [144] found that transduction rates were directly related to the recipient cell concentration. Prokaryotic cells are typically unevenly distributed throughout freshwater lacustrine systems. In the bulk water phase, prokaryotic cell concentrations can vary over space and time [145] but are typically relatively low, resulting in substantial spatial isolation that reduces the probability of HGT. Local regions of relatively high cell density include physical interfaces within the water column (e.g. boundaries of the metalimnion), the air-water and sediment-water interfaces, and biofilms both suspended within the water column (e.g. lake snow) and present at abiotic and biotic solid surfaces. Bacterial and viral abundance and production have been found to vary significantly among strata in stratified water columns, but consistently appear to be elevated in the metalimnion [146-148], indicating this region is a hotspot for HGT and that rates of gene transfer likely differ among epilimnetic, metalimnetic and hypolimnetic regions. A greater number of transconjugants was detected at the air-water interface relative to the bulk underlying water in a model freshwater system, suggesting that the surface microlayer represents a distinct microzone for HGT in freshwater lakes [29].
In aquatic environments, prokaryotes overwhelmingly tend to form and be found in biofilms at surfaces [149]. As these structures maintain a high density of metabolically active cells surrounded by a matrix containing potentially transformable extracellular DNA and capable of facilitating extracellular communication, providing protection from predation, and trapping nutrients and other chemicals, they are potential hotspots for HGT [150]. Underscoring the prevalence of HGT in biofilm settings, conjugation machinery [151], conjugative plasmids [152], and phage particles [153] have important roles in biofilm formation. Increased rates of HGT have been observed in biofilms relative to planktonic cells [154-157]. In a microcosm study designed to mimic a marine environment, conjugation frequencies were typically higher in biofilms compared to the bulk water phase, particularly at the biofilm surface in contact with the surrounding environment [158]. Biofilm stability and the stage of biofilm development may influence HGT. Regular disturbance of the spatial organization of prokaryotic cells in an in vitro biofilm strongly improved plasmid invasiveness, suggesting that the periodic breakup of biofilms (e.g. due to wind/solar driven turbulence) may promote HGT [157]. In cultured Escherichia coli and Pseudomonas aeruginosa mixed-biofilm communities, phage particles were observed by transmission electron microscopy to be closely associated with the biofilm matrix [159]. Following their release from lysogenic Staphylococcus aureus cells, phage particles remained detectable for longer periods in biofilm cultures than in planktonic cultures due to protection from degradation [160], suggesting that transduction may be more prevalent in freshwater biofilms relative to the surrounding bulk water phase.
Within biofilms, microscale gradients in oxygen, pH, and concentrations of nutrients, electron donors and acceptors, signalling molecules, trace elements, and antibiotics can produce microenvironments with distinct biogeochemistry [161-164] that may differentially influence local rates of HGT. Sites of biofilm formation and potential HGT hotspots in freshwater lakes include suspended particulate matter, submerged bulk abiotic surfaces (e.g. rocks) and biotic surfaces (e.g. epilithon of aquatic plants), and the internal structures of aquatic plants and animals (e.g. digestive tract of filter feeding bivalves) [23,165] (Figure 1(b)). As noted by Hermansson and Linberg [166] and Hill and Top [124], rates of HGT likely differ substantially among the numerous prokaryotic biofilms present in a lacustrine system, reflecting in part the highly variable environmental conditions that are present.
3.9. Nutrient Availability
The availability of nutrients for prokaryotic cell growth is potentially an important determinant of HGT rates in freshwater lakes. In vitro conjugation rates in pure cultures of heterotrophic bacteria have been positively correlated with the availability of rich nutrient sources containing phosphorus, nitrogen, and organic carbon [30,67], as well as with organic carbon alone [157,167]. Additionally, conjugation rates between introduced heterotrophic bacteria were found to be significantly higher in sterile nutrient-rich sewage water compared to sterile lake water [168]. Interestingly, the positive relationship between nutrient availability and conjugative plasmid transfer appears to be attenuated when nutrient concentrations rise above a particular threshold value in vitro [169,170], suggesting that a similar threshold may exist in natural environments. The latency period between phage infection and cell lysis, as well as the resulting number of released phage particles, are reduced in starved bacterial cells compared to actively metabolizing cells, indicating that transduction rates are reduced under conditions of nutrient limitation [171]. In addition to serving as a vector for transformation, extracellular DNA is a source of exogenous nucleotides, labile carbon, nitrogen, and phosphorus for microbial communities [119]. Reduced availability of these nutrients may increase the utilization of extracellular DNA by prokaryotes as a nutrient source may increase, potentially reducing the amount of transformable DNA available for HGT [172]. Collectively, these findings suggest that HGT rates in freshwater lacustrine systems are directly related to the availability of nutrients for prokaryotic growth and that HGT rates in heterotrophic prokaryotes are likely to be higher in nutrient/organic carbon rich systems. In freshwater lakes, bacterial production and growth rates can be constrained by the availability of nitrogen, phosphorus, and/or organic carbon [173], with phosphorus most commonly being the limiting nutrient [174]. It is important to note that most investigations of the effects of nutrient availability on HGT employed a single chemoorganoheterotrophic species of bacteria to assess the effects of organic carbon or general nutrient limitation, which precluded the investigation of other heterotrophic and autotrophic prokaryotes as well as any specific effects of phosphorus and nitrogen limitation on HGT in freshwater prokaryotic communities.
Nutrient limitation can also stimulate HGT, blurring the relationship between nutrient availability and HGT that has been established in vitro and exemplifying the likely complexity of HGT regulation within natural environments. As with DNA damage resulting from exposure to genotoxic chemicals or ultraviolet radiation, starvation can lead to the activation of bacterial stress responses that in turn can promote HGT. Amino acid and thymine deprivation can induce prophage and thereby promote transduction [175]. The development of competence in vitro has been observed in cells in association with a reduction in metabolic activity, which can result from nutrient limitation [50,176]. Conjugation has been demonstrated in laboratory seawater microcosms even after starvation for up to 15 days [177], indicating that nutrient limitation does not preclude the potential for conjugation in a freshwater prokaryotic community. Hausner and Wuertz [154] found that conjugation rates were not appreciably diminished by nutrient limitation under laboratory conditions in bacterial biofilms, and suggested that although conjugation requires energy, starved cells that constitutively express their conjugation apparatus are still able to participate in HGT.
Viral infection rates and bacterial abundance and metabolic activity are generally higher in nutrient-rich freshwater systems [36,178], indicating greater rates of HGT. Within lacustrine systems, HGT rates may reflect spatial and temporal variations in nutrient availability. Epilimnetic waters of highly productive stratified lakes are typically depleted of nutrients during summer stratification as organic matter settles out of this compartment into the underlying metalimnion and hypolimnion [179]. The availability of phosphorus, nitrogen, and organic carbon is generally increased during turnover events, storm events, and the spring melting of snow and ice in temperate climates, as nutrient-rich material is introduced or redistributed within a system. Nutrient inputs may be particularly high in urban and agriculturally impacted areas, and point sources within lakes (e.g. wastewater treatment plant effluent) may create localized HGT hotspots. Concentrations of organic and inorganic nutrients tend to be greater in interfacial regions including the metalimnion, air-water interface [128,180], and sedimentwater interface [181] compared to the bulk water column. Further, biofilms, commonly observed at such interfaces, can sorb and accumulate nutrients from the surrounding environment and the extracellular polymeric substances of which they are composed can serve as a nutrient source (e.g. extracellular DNA) [47].
3.10. Ecological Interactions
In freshwater lacustrine environments, interactions within prokaryotic communities and among prokaryotes and plants, algae, and protozoa can potentially influence rates of prokaryotic HGT. Bacterial quorum sensing (intercellular communication) systems can regulate conjugative transfer [182,183], prophage induction [184], and transformation [185]. Further, addition of Escherichia coli cell-free supernatant was observed to increase the frequency of transformation in Bacillus subtilis cells nearly ten-fold, suggesting that organic factors produced by one species in a freshwater prokaryotic community can stimulate transformation in a distantly related species [138]. Investigations of HGT to date have largely utilized single bacterial species, such that the influence of interacttions between different prokaryotic taxa on gene transfer remains poorly defined.
The rhizosphere of terrestrial plants has been identified as a hotspot for HGT, in part due to the availability of nutrient-rich plant root exudates [124,186]. Similarly, in lacustrine systems, aquatic plants and algae may enhance HGT in nearby prokaryotic communities. Conjugative plasmid transfer between bacteria in a microcosm was enhanced in the presence of the aquatic marsh plant Echinochlora crusgalli [187]. The presence of algae or algal exudates can enhance in vitro rates of conjugation [188] and transformation [189] in prokaryotes, suggesting that algal blooms may be hotspots for HGT. Under laboratory conditions, cocultivation of Bacillus subtilis with the ciliate protozoa Tetrahymena thermophila or metabolites from co-cultures decreased transformation rates, with the inhibitory effect of the metabolites being attenuated when the photosynthetic alga Euglena gracilis was present in culture [190]. In contrast, the presence of ciliates, which are ubiquitous in freshwater lakes, increased conjugation rates in cultures of Escherichia coli by facilitating contact between donor and recipient strains through co-accumulation in its vesicles [191]. Intercellular communication between bacteria is enhanced in biofilms [192] and freshwater biofilm communities typically include prokaryotes, algae, and protozoa, suggesting that they are important sites for ecological regulation of HGT in freshwater lacustrine systems.
More generally, low molecular weight organic acids (e.g. tartrate, citrate) derived from organic matter and associated with regions of high biological activity can compete with DNA for adsorption to cells, potentially inhibiting transformation [193]. Chitin, a biopolymer found in fungi, insects, molluscs, and crustaceans in aquatic environments, has been shown to induce competence in bacteria capable of colonizing and degrading it in vitro [194]. This suggests that transformation rates may be influenced by the distribution and abundance of chitin producing organisms within freshwater lakes.
3.11. Prokaryotic Community Composition
Generally speaking, HGT in the majority of freshwater prokaryotes remains uncharacterized and taxon-specific variables that may influence HGT remain to be determined. However, several lines of evidence suggest that the composition of freshwater prokaryotic communities influences the character of HGT taking place within them, although much more research is necessary to firmly establish such a relationship. First, metabolisms limited to particular taxa, as discussed previously, can differentially alter local biogeochemical conditions, in turn potentially influencing HGT among impacted prokaryotes. Second, the formation of consortia dependent on the presence of particular community members may impact HGT rates through changes in donor and recipient cell numbers, metabolic activities, and proximity [124]. For example, chemoorganoheterotrophic bacteria tend to accumulate around cyanobacterial heterocysts, taking advantage of secreted organic carbon and nutrients [195]. Third, plasmids and phages can have specific host ranges [196] and transforming DNA can be tagged such that it will be preferentially taken up by specific taxa [197], such that the extent to which they mediate HGT within a prokaryotic community is dependent on the presence and relative abundance of particular taxa. High prokaryotic diversity can potentially inhibit transduction rates via cells not susceptible to infection by a particular phage interfering with initial virus-host contact [37]. Fourth, nuclease activity and DNA release may differ depending on prokaryotic community composition, influencing transformation rates via the availability of transformable DNA [176]. Finally, as discussed previously, communication within prokaryotic communities via extracellular signalling molecules with taxa-specific production and responses can differentially regulate HGT.
Prokaryotic community composition can differ substantially between freshwater lakes [198,199] and within systems can vary with depth and site location, as well as over seasonal and annual timescales [200-203]. Further, community composition can vary between epilimnetic and hypolimnetic regions within summer stratified lakes [204-206] and planktonic bacterial communities can differ from those associated with suspended particulate matter [207,208]. Finally, regular and episodic system perturbations (e.g. seasonal lake mixing events, addition of prokaryotes and nutrients via wastewater input) may appreciably alter community composition [206,209]. These spatial and temporal variations in lacustrine prokaryotic community composition may be associated with different HGT rates and participants.
4. Concluding Remarks
Although the three principal mechanisms of HGT have been demonstrated in freshwater environments, field studies linking HGT to specific biogeochemical parameters are relatively scarce and experimental studies assessing natural prokaryotic communities are equally rare. These linkages, although established in many cases using laboratory-based approaches with chemoorganoheterotrophic organisms, have yet to be confirmed in freshwater lacustrine systems. Consequently, how and to what extent individual biogeochemical factors influence HGT in freshwater lakes remains poorly understood. An appreciation for the substantial spatial and temporal variability in factors capable of influencing HGT within freshwater lakes is necessary to fully characterize environmental gene transfer. In particular, interfacial regions, whether between thermally stratified zones of water, between bottom sediments and overlying water, or between the water surface and the atmosphere, are areas that should be targeted for further investigation of freshwater lacustrine HGT. Further, greater exploration of HGT within autotrophic guilds, a key component of environmental prokaryotic communities, is required to fully constrain these processes within freshwater systems. Culture-independent molecular techniques (e.g. [27,210,211]) provide a means of in situ assessment of HGT in freshwater prokaryotic communities. It is anticipated that the coupling of these techniques with biogeochemical characterization will further understanding of HGT occurrence, controls and impacts within freshwater lakes.
REFERENCES
- E. V. Koonin and Y. I. Wolf, “Genomics of Bacteria and Archaea: The Emerging Dynamic View of the Prokaryotic World,” Nucleic Acids Research, Vol. 36, No. 21, 2008, pp. 6688-6719. doi:10.1093/nar/gkn668
- O. Zhaxybayeva and W. F. Doolittle, “Lateral Gene Transfer,” Current Biology, Vol. 21, No. 7, 2011, pp. R242-R246. doi:10.1016/j.cub.2011.01.045
- A. O. Summers, “Genetic Linkage and Horizontal Gene Transfer, the Roots of the Antibiotic Multi-Resistance Problem,” Animal Biotechnology, Vol. 17, No. 2, 2006, pp. 125-135. doi:10.1080/10495390600957217
- D. Springael and E. M. Top, “Horizontal Gene Transfer and Microbial Adaptation to Xenobiotics: New Types of Mobile Genetic Elements and Lessons from Ecological Studies,” Trends in Microbiology, Vol. 12, No. 2, 2004, pp. 53-58. doi:10.1016/j.tim.2003.12.010
- B. Liang, G. Wang, Y. Zhao, K. Chen, S. Li and J. Jiang, “Facilitation of Bacterial Adaptation to ChlorothalonilContaminated Sites by Horizontal Transfer of the Chlorothalonil Hydrolytic Dehalogenase Gene,” Applied and Environmental Microbiology, Vol. 77, No. 12, 2011, pp. 4268- 4272. doi:10.1128/AEM.02457-10
- J. M. Monier, D. Bernillon, E. Kay, A. Faugier, O. Rybalka, Y. Dessaux, P. Simonet and T. M. Vogel, “Detection of Potential Transgenic Plant DNA Recipients among Soil Bacteria,” Environmental Biosafety Research, Vol. 6, No. 1-2, 2007, pp. 71-83. doi:10.1051/ebr:2007036
- F. Donnarumma, D. Paffetti, G. Stotzky, R. Giannini and C. Vettori, “Potential Gene Exchange between Bacillus thuringiensis subsp. kurstaki and Bacillus spp. in Soil in Situ,” Soil Biology and Biochemistry, Vol. 42, No. 8, 2010, pp. 1329-1337. doi:10.1016/j.soilbio.2010.03.014
- J. B. Cotner and B. A. Biddanda, “Small Players, Large Role: Microbial Influence on Biogeochemical Processes in Pelagic Aquatic Ecosystems,” Ecosystems, Vol. 5, No. 2, 2002, pp. 105-121. doi:10.1007/s10021-001-0059-3
- US Environmental Protection Agency, “Great Lakes,” 2011. http://www.epa.gov/greatlakes/statsrefs.html
- A. Pruden, R. Pei, H. Storteboom and K. H. Carlson, “Antibiotic Resistance Genes as Emerging Contaminants: Studies in Northern Colorado,” Environmental Science and Technology, Vol. 40, No. 23, 2006, pp. 7445-7450. doi:10.1021/es060413l
- J. L. Martinez, “Environmental Pollution by Antibiotics and by Antibiotic Resistance Determinants,” Environmental Pollution, Vol. 157, No. 11, 2009, pp. 2893-2902. doi:10.1016/j.envpol.2009.05.051
- H. K. Allen, J. Donato, H. H. Wang, K. A. Cloud-Hansen, J. Davies and J. Handelsman, “Call of the Wild: Antibiotic Resistance Genes in Natural Environments,” Nature Reviews Microbiology, Vol. 8, No. 4, 2010, pp. 251-259. doi:10.1038/nrmicro2312
- N. G. Taylor, D. W. Verner-Jeffreys and C. Baker-Austin, “Aquatic Systems: Maintaining, Mixing and Mobilising Antimicrobial Resistance?” Trends in Ecology and Evolution, Vol. 26, No. 6, 2011, pp. 278-284. doi:10.1016/j.tree.2011.03.004
- A. Lupo, S. Coyne and T. U. Berendonk, “Origin and Evolution of Antibiotic Resistance: The Common Mechanisms of Emergence and Spread in Water Bodies,” Frontiers in Microbiology, Vol. 3, 2012, p. 18. doi:10.3389/fmicb.2012.00018
- L. S. Frost and G. Koraimann, “Regulation of Bacterial Conjugation: Balancing Opportunity with Adversity,” Future Microbiology, Vol. 5, No. 7, 2010, pp. 1057-1071. doi:10.2217/fmb.10.70
- O. Johnsborg, V. Eldholm and L. S. Håvarstein, “Natural Genetic Transformation: Prevalence, Mechanisms and Function,” Research in Microbiology, Vol. 158, No. 10, 2007, pp. 767-778. doi:10.1016/j.resmic.2007.09.004
- C. Canchaya, G. Fournous, S. Chibani-Chennoufi, M. L. Dillmann and H. Brüssow, “Phage as Agents of Lateral Gene Transfer,” Current Opinion in Microbiology, Vol. 6, No. 4, 2003, pp. 417-424. doi:10.1016/S1369-5274(03)00086-9
- M. Martinez-Garcia, B. K. Swan, N. J. Poulton, M. L. Gomez, D. Masland, M. E. Sieracki and R. Stepanauskas, “High-Throughput Single-Cell Sequencing Identifies Photoheterotrophs and Chemoautotrophs in Freshwater Bacterioplankton,” The ISME Journal, Vol. 6, No. 1, 2012, pp. 113-123. doi:10.1038/ismej.2011.84
- D. J. Saye, O. Ogunseitan, G. S. Sayler and R. V. Miller, “Potential for Transduction of Plasmids in a Natural Freshwater Environment: Effect of Plasmid Donor Concentration and a Natural Microbial Community on Transduction in Pseudomonas aeruginosa,” Applied and Environmental Microbiology, Vol. 53, No. 5, 1987, pp. 987- 995.
- S. B. O’Morchoe, O. Ogunseitan, G. S. Sayler and R. V. Miller, “Conjugal Transfer of R68.45 and FP5 between Pseudomonas aeruginosa Strains in a Freshwater Environment,” Applied and Environmental Microbiology, Vol. 54, No. 8, 1988, pp. 1923-1929.
- H. G. Williams, M. J. Day, J. C. Fry and G. J. Stewart, “Natural Transformation in River Epilithon,” Applied and Environmental Microbiology, Vol. 62, No. 8, 1996, pp. 2994-2998.
- A. Caro-Quintero, J. Deng, J. Auchtung, I. Brettar, M. G. Höfle, J. Klappenbach and K. T. Konstantinidis, “Unprecedented Levels of Horizontal Gene Transfer among Spatially Co-Occurring Shewanella Bacteria from the Baltic Sea,” The ISME Journal, Vol. 5, No. 1, 2011, pp. 131-140. doi:10.1038/ismej.2010.93
- K. E. Ashelford, J. C. Fry, M. J. Day, K. E. Hill, M. A. Learner, J. R. Marchesi, C. D. Perkins and A. J. Weightman, “Using Microcosms to Study Gene Transfer in Aquatic Habitats,” FEMS Microbiology Ecology, Vol. 23, No. 2, 1997, pp. 81-94. doi:10.1111/j.1574-6941.1997.tb00393.x
- J. D. van Elsas, J. Fry, P. Hirsch and S. Molin, “Ecology of Plasmid Transfer and Spread,” In: C. M. Thomas, Ed., The Horizontal Gene Pool, Harwood, Amsterdam, 2000, pp. 175-206. doi:10.4324/9780203304334_chapter_4
- M. Shintani, N. Fukushima, M. Tezuka, H. Yamane and H. Nojiri, “Conjugative Transfer of the IncP-7 Carbazole Degradative Plasmid, pCAR1, in River Water Samples,” Biotechnology Letters, Vol. 30, No. 1, 2008, pp. 117-122. doi:10.1007/s10529-007-9519-y
- T. Kenzaka, K. Tani and M. Nasu, “High-Frequency Phage-Mediated Gene Transfer in Freshwater Environments Determined at Single-Cell Level,” The ISME Journal, Vol. 4, No. 5, 2010, pp. 648-659. doi:10.1038/ismej.2009.145
- F. Maruyama, K. Tani, T. Kenzaka, N. Yamaguchi and M. Nasu, “Quantitative Determination of Free-DNA Uptake in River Bacteria at the Single-Cell Level by in Situ Rolling-Circle Amplification,” Applied and Environmental Microbiology, Vol. 72, No. 9, 2006, pp. 6248-6256. doi:10.1128/AEM.03035-05
- F. Maruyama, K. Tani, T. Kenzaka, N. Yamaguchi and M. Nasu, “Application of Real-Time Long and Short Polymerase Chain Reaction for Sensitive Monitoring of the Fate of Extracellular Plasmid DNA Introduced into River Waters,” Microbes and Environments, Vol. 23, No. 3, 2008, pp. 229-236. doi:10.1264/jsme2.23.229
- G. W. Jones, L. Baines and F. J. Genthner, “Heterotrophic Bacteria of the Freshwater Neuston and Their Ability to Act as Plasmid Recipients under Nutrient Deprived Conditions,” Microbial Ecology, Vol. 22, No. 1, 1991, pp. 15- 25. doi:10.1007/BF02540210
- M. J. Bale, J. C. Fry and M. J. Day, “Plasmid Transfer between Strains of Pseudomonas aeruginosa on Membrane Filters Attached to River Stones,” Journal of General Microbiology, Vol. 133, No. 11, 1987, pp. 3099- 3107.
- M. J. Bale, M. J. Day and J. C. Fry, “Novel Method for Studying Plasmid Transfer in Undisturbed River Epilithon,” Applied and Environmental Microbiology, Vol. 54, No. 11, 1988, pp. 2756-2758.
- M. J. Bale, J. C. Fry and M. J. Day, “Transfer and Occurrence of Large Mercury Resistance Plasmids in River Epilithon,” Applied and Environmental Microbiology, Vol. 54, No. 4, 1988, pp. 972-978.
- M. G. Jobling, S. E. Peters and D. A. Ritchie, “Plasmid-Borne Mercury Resistance in Aquatic Bacteria,” FEMS Microbiology Letters, Vol. 49, No. 1, 1988, pp. 31-37.
- J. C. Fry and M. J. Day, “Plasmid Transfer in the Epilithon,” In: J. C. Fry and M. J. Day, Eds., Bacterial Genetics in Natural Environments, Chapman and Hall, London, 1990, pp. 172-181. doi:10.1007/978-94-009-1834-4_5
- M. G. Weinbauer, “Ecology of Prokaryotic Viruses,” FEMS Microbiology Reviews, Vol. 28, No. 2, 2004, pp. 127-181. doi:10.1016/j.femsre.2003.08.001
- K. E. Wommack and R. R. Colwell, “Virioplankton: Viruses in Aquatic Ecosystems,” Microbiology and Molecular Biology Reviews, Vol. 64, No. 1, 2000, pp. 69-114. doi:10.1128/MMBR.64.1.69-114.2000
- R. V. Miller, “Environmental Bacteriophage-Host Interactions: Factors Contribution to Natural Transduction,” Antonie Van Leeuwenhoek, Vol. 79, No. 2, 2001, pp. 141- 147. doi:10.1023/A:1010278628468
- W. D. Morrison, R. V. Miller and G. S. Sayler, “Frequency of F116-Mediated Transduction of Pseudomonas aeruginosa in a Freshwater Environment,” Applied and Environmental Microbiology, Vol. 36, No. 5, 1978, pp. 724-730.
- D. J. Saye, O. Ogunseitan, G. S. Sayler and R. V. Miller, “Transduction of Linked Chromosomal Genes between Pseudomonas aeruginosa Strains during Incubation in Situ in a Freshwater Habitat,” Applied and Environmental Microbiology, Vol. 56, No. 1, 1990, pp. 140-145.
- S. Ripp and R. V. Miller, “Effects of Suspended Particulates on the Frequency of Transduction among Pseudomonas aeruginosa in a Freshwater Environment,” Applied and Environmental Microbiology, Vol. 61, No. 4, 1995, pp. 1214-1219.
- M. K. Amin and M. J. Day, “Donor and Recipient Effects on Transduction Frequency in Situ,” REGEM 1 Programme for the 1st International Conference on the Release of Genetically Engineered Microorganisms, Cardiff, 5-8 April 1988, p. 11.
- K. M. Nielsen, P. J. Johnsen, D. Bensasson and D. Daffonchio, “Release and Persistence of Extracellular DNA in the Environment,” Environmental Biosafety Research, Vol. 6, No. 1-2, 2007, pp. 37-53. doi:10.1051/ebr:2007031
- D. M. Karl and M. D. Bailiff, “The Measurement and Distribution of Dissolved Nucleic Acids in Aquatic Environments,” Limnology and Oceanography, Vol. 34, No. 3, 1989, pp. 543-558. doi:10.4319/lo.1989.34.3.0543
- J. H. Paul, S. C. Jiang and J. B. Rose, “Concentration of Viruses and Dissolved DNA from Aquatic Environments by Vortex Flow Filtration,” Applied and Environmental Microbiology, Vol. 57, No. 8, 1991, pp. 2197-2204.
- W. Siuda, R. J. Chróst and H. Güde, “Distribution and Origin of Dissolved DNA in Lakes of Different Trophic States,” Aquatic Microbial Ecology, Vol. 15, No. 1, 1998, pp. 89-96. doi:10.3354/ame015089
- S. C. Jiang and J. H. Paul, “Viral Contribution to Dissolved DNA in the Marine Kingdom as Determined by Differential Centrifugation and Kingdom Probing,” Applied and Environmental Microbiology, Vol. 61, No. 1, 1995, pp. 317-325.
- H. C. Flemming and J. Wingender, “The Biofilm Matrix,” Nature Reviews Microbiology, Vol. 8, No. 9, 2010, pp. 623-633. doi:10.1038/nrmicro2415
- T. Hara and S. Ueda, “A Study on the Mechanism of DNA Excretion from P. aeruginosa KYU-1. Effect of Mitomycin C on Extracellular DNA Production,” Agricultural Chemistry and Biotechnology, Vol. 45, No. 11, 1981, pp. 2457-2461. doi:10.1271/bbb1961.45.2457
- M. G. Lorenz, D. Gerjets and W. Wackernagel, “Release of Transforming Plasmid and Chromosomal DNA from Two Cultured Soil Bacteria,” Archives of Microbiology, Vol. 156, No. 4, 1991, pp. 319-326. doi:10.1007/BF00263005
- G. J. Stewart, “Transformation in Natural Environments,” In: E. M. H. Wellington and J. D. van Elsas, Eds., Genetic Interactions among Microorganisms in the Natural Environment, Pergamon Press, Oxford, 1992, pp. 216-234.
- G. Romanowski, M. G. Lorenz and W. Wackernagel, “Adsorption of Plasmid DNA to Mineral Surfaces and Protection against DNase I,” Applied and Environmental Microbiology, Vol. 57, No. 4, 1991, pp. 1057-1061.
- V. Torsvik, L. Øvreås and T. F. Thingstad, “Prokaryotic Diversity—Magnitude, Dynamics, and Controlling Factors,” Science, Vol. 296, No. 5570, 2002, pp. 1064-1066. doi:10.1126/science.1071698
- E. J. Biers, S. Sun and E. C. Howard, “Prokaryotic Genomes and Diversity in Surface Ocean Waters: Interrogating the Global Ocean Sampling Metagenome,” Applied and Environmental Microbiology, Vol. 75, No. 7, 2009, pp. 2221-2229. doi:10.1128/AEM.02118-08
- R. J. Newton, S. E. Jones, A. Eiler, K. D. McMahon and S. Bertilsson, “A Guide to the Natural History of Freshwater Lake Bacteria,” Microbiology and Molecular Biology Reviews, Vol. 75, No. 1, 2011, pp. 14-49. doi:10.1128/MMBR.00028-10
- L. A. Warren and E. A. Haack, “Biogeochemical Controls on Metal Behaviour in Freshwater Environments,” EarthScience Reviews, Vol. 54, No. 4, 2001, pp. 261-320. doi:10.1016/S0012-8252(01)00032-0
- A. Bissett, A. Reimer, D. de Beer, F. Shiraishi and G. Arp, “Metabolic Microenvironmental Control by Photosynthetic Biofilms under Changing Macroenvironmental Temperature and pH Conditions,” Applied and Environmental Microbiology, Vol. 74, No. 20, 2008, pp. 6306-6312. doi:10.1128/AEM.00877-08
- D. A. Fike, C. L. Gammon, W. Ziebis and V. J. Orphan, “Micron-Scale Mapping of Sulfur Cycling across the Oxycline of a Cyanobacterial Mat: A Paired nanoSIMS and CARD-FISH Approach,” The ISME Journal, Vol. 2, No. 7, 2008, pp. 749-759. doi:10.1038/ismej.2008.39
- T. Borch, R. Kretzschmar, A. Kappler, P. Van Cappellen, M. Ginder-Vogel, A. Voegelin and K. Campbell, “Biogeochemical Redox Processes and Their Impact on Contaminant Dynamics,” Environmental Science and Technology, Vol. 44, No. 1, 2010, pp. 15-23. doi:10.1021/es9026248
- T. Lawley, B. M. Wilkins and L. S. Frost, “Bacterial Conjugation in Gram-Negative Bacteria,” In: B. E. Funnell and G. J. Philips, Eds., Plasmid Biology, ASM Press, Washington DC, 2004, pp. 203-226.
- [61] G. Alonso, K. Baptista, T. Ngo and D. E. Taylor, “Transcriptional Organization of the Temperature-Sensitive Transfer System from the IncHI1 Plasmid R27,” Microbiology, Vol. 151, No. 11, 2005, pp. 3563-3573. doi:10.1099/mic.0.28256-0
- [62] F. W. Shiah and H. W. Ducklow, “Temperature Regulation of Heterotrophic Bacterioplankton Abundance, Production, and Specific Growth Rate in Chesapeake Bay,” Limnology and Oceanography, Vol. 39, No. 6, 1994, pp. 1243-1258. doi:10.4319/lo.1994.39.6.1243
- [63] A. S. P. Ram, D. Boucher, T. Sime-Ngando, D. Debroas and J. C. Romagoux, “Phage Bacteriolysis, Protistan Bacterivory Potential, and Bacterial Production in a Freshwater Reservoir: Coupling with Temperature,” Microbial Ecology, Vol. 50, No. 1, 2005, pp. 64-72. doi:10.1007/s00248-004-0110-y
- [64] W. F. Yue, M. Du and M. J. Zhu, “High Temperature in Combination with UV Irradiation Enhances Horizontal Transfer of stx2 Gene from E. coli O157:H7 to NonPathogenic E. coli,” PLoS ONE, Vol. 7, No. 2, 2012, Article ID: e31308. doi:10.1371/journal.pone.0031308
- [65] I. Ahrenholtz, M. G. Lorenz and W. Wackernagel, “The Extracellular Nuclease of Serratia marcescens: Studies on the Activity in Vitro and Effect on Transforming DNA in a Groundwater Aquifer Microcosm,” Archives of Microbiology, Vol. 161, No. 2, 1994, pp. 176-183.
- [66] K. Matsui, M. Honjo and Z. Kawabata, “Estimation of the Fate of Dissolved DNA in Thermally Stratified Lake Water from the Stability of Exogenous Plasmid DNA,” Aquatic Microbial Ecology, Vol. 26, No. 1, 2001, pp. 95- 102. doi:10.3354/ame026095
- [67] I. Auzat, S. Chapuy-Regaud, G. Le Bras, D. D. Santos, A. D. Ogunniyi, I. L. Thomas, J. R. Garel, J. C. Paton and M. C. Trombe, “The NADH Oxidase of Streptococcus pneumoniae: Its Involvement in Competence and Virulence,” Molecular Microbiology, Vol. 34, No. 5, 1999, pp. 1018- 1028. doi:10.1046/j.1365-2958.1999.01663.x
- [68] D. R. Stallions and R. Curtiss, “Bacterial Conjugation under Anaerobic Conditions,” Journal of Bacteriology, Vol. 111, No. 1, 1972, pp. 294-295.
- [69] J. E. Król, H. D. Nguyen, L. M. Rogers, H. Beyenal, S. M. Krone and E. M. Top, “Increased Transfer of a Multidrug Resistance Plasmid in Escherichia coli Biofilms at the Air-Liquid Interface,” Applied and Environmental Microbiology, Vol. 77, No. 15, 2011, pp. 5079-5088. doi:10.1128/AEM.00090-11
- [70] L. S. Frost and J. Simon, “Studies on the Pili of the Promiscuous Plasmid RP4,” In: C. I. Kado and J. H. Crosa, Eds., Molecular Mechanisms of Bacterial Virulence, Kluwer Academic Publishers, Dordrecht, 1994, pp. 47-65. doi:10.1007/978-94-011-0746-4_4
- [71] P. Viljanen and J. Boratynski, “The Susceptibility of Conjugative Resistance Transfer in Gram-Negative Bacteria to Physicochemical and Biochemical Agents,” FEMS Microbiology Letters, Vol. 88, No. 1, 1991, pp. 43-54. doi:10.1111/j.1574-6968.1991.tb04956.x
- [72] C. A. Pinedo and B. F. Smets, “Conjugal TOL Transfer from Pseudomonas putida to Pseudomonas aeruginosa: Effects of Restriction Proficiency, Toxicant Exposure, Cell Density Ratios, and Conjugation Detection Method on Observed Transfer Efficiencies,” Applied and Environmental Microbiology, Vol. 71, No. 1, 2005, pp. 51-57. doi:10.1128/AEM.71.1.51-57.2005
- [73] A. Schäfer, J. Kalinowski and A. Pühler, “Increased Fertility of Corynebacterium glutamicum Recipients in Intergeneric Matings with Escherichia coli after Stress Exposure,” Applied and Environmental Microbiology, Vol. 60, No. 2, 1994, pp. 756-759.
- [74] R. A. Edwards, R. A. Helm and S. R. Maloy, “Increasing DNA Transfer Efficiency by Temporary Inactivation of Host Restriction,” Biotechniques, Vol. 26, No. 5, 1999, pp. 892-900.
- [75] J. P. Claverys, M. Prudhomme and B. Martin, “Induction of Competence Regulons as a General Response to Stress in Gram-Positive Bacteria,” Annual Review of Microbiology, Vol. 60, 2006, pp. 451-475. doi:10.1146/annurev.micro.60.080805.142139
- [76] M. Prudhomme, L. Attaiech, G. Sanchez, B. Martin and J. P. Claverys, “Antibiotic Stress Induces Genetic Transformability in the Human Pathogen Streptococcus pneumonia,” Science, Vol. 313, No. 5783, 2006, pp. 89-92. doi:10.1126/science.1127912
- [77] C. Úbeda, E. Maiques, E. Knecht, Í. Lasa, R. P. Novick and J. R. Penadés, “Antibiotic-Induced SOS Response Promotes Horizontal Dissemination of Pathogenicity Island-Encoded Virulence Factors in Staphylococci,” Molecular Microbiology, Vol. 56, No. 3, 2005, pp. 836-844. doi:10.1111/j.1365-2958.2005.04584.x
- [78] E. Maiques, C. Úbeda, S. Campoy, N. Salvador, Í. Lasa, R. P. Novick, J. Barbé and J. R. Penadés, “β-Lactam Antibiotics Induce the SOS Response and Horizontal Transfer of Virulence Factors in Staphylococcus aureus,” Journal of Bacteriology, Vol. 188, No. 7, 2006, pp. 2726- 2729. doi:10.1128/JB.188.7.2726-2729.2006
- [79] J. W. Beaber, B. Hochhut and M. K. Waldor, “SOS Response Promotes Horizontal Dissemination of Antibiotic Resistance Genes,” Nature, Vol. 427, No. 6969, 2004, pp. 72-74. doi:10.1038/nature02241
- [80] T. G. Rossman, M. Molina and L. W. Meyer, “The Genetic Toxicology of Metal Compounds: I. Induction of λ Prophage in E. coli WP2S (λ),” Environmental Mutagenesis, Vol. 6, No. 1, 1984, pp. 59-69. doi:10.1002/em.2860060108
- [81] T. G. Rossman, M. Molina, L. Meyer, P. Boone, C. B. Klein, Z. Wang, F. Li, W. C. Lin and P. L. Kinney, “Performance of 133 Compounds in the Lambda Prophage Induction Endpoint of the Microscreen Assay and a Comparison with S. typhimurium Mutagenicity and Rodent Carcinogenicity Assays,” Mutation Research, Vol. 260, No. 4, 1991, pp. 349-367. doi:10.1016/0165-1218(91)90021-D
- [82] D. M. DeMarini and H. G. Brooks, “Induction of Prophage Lambda by Chlorinated Organics: Detection of Some Single-Species/Single-Site Carcinogens,” Environmental and Molecular Mutagenesis, Vol. 19, No. 2, 1992, pp. 98-111. doi:10.1002/em.2850190204
- [83] N. Soberón, R. Martín and J. E. Suárez, “New Method for Evaluation of Genotoxicity, Based on the Use of RealTime PCR and Lysogenic Gram-Positive and GramNegative Bacteria,” Applied and Environmental Microbiology, Vol. 73, No. 9, 2007, pp. 2815-2819. doi:10.1128/AEM.00407-07
- [84] L. R. Mesak, V. Miao and J. Davies, “Effects of Subinhibitory Concentrations of Antibiotics on SOS and DNA Repair Gene Expression in Staphylococcus aureus,” Antimicrobial Agents and Chemotherapy, Vol. 52, No. 9, 2008, pp. 3394-3397. doi:10.1128/AAC.01599-07
- [85] Z. Baharoglu and D. Mazel, “Vibrio cholerae Triggers SOS and Mutagenesis in Response to a Wide Range of Antibiotics: A Route towards Multiresistance,” Antimicrobial Agents and Chemotherapy, Vol. 55, No. 5, 2011, pp. 2438-2441. doi:10.1128/AAC.01549-10
- [86] V. M. F. Vargas, S. B. Migliavacca, A. C. de Melo, R. C. Horn, R. R. Guidobono, I. C. F. de Sá Ferreira and M. H. D. Pestana, “Genotoxicity Assessment in Aquatic Environments under the Influence of Heavy Metals and Organic Contaminants,” Mutation Research, Vol. 490, No. 2, 2001, pp. 141-158. doi:10.1016/S1383-5718(00)00159-5
- [87] E. Kristiansson, J. Fick, A. Janzon, R. Grabic, C. Rutgersson, B. Weijdegård, H. Söderström and D. G. J. Larsson, “Pyrosequencing of Antibiotic-Contaminated River Sediments Reveals High Levels of Resistance and Gene Transfer Elements,” PLoS ONE, Vol. 6, No. 2, 2011, e17038. doi:10.1371/journal.pone.0017038
- [88] K. Smalla, A. S. Haines, K. Jones, E. Krögerrecklenfort, H. Heuer, M. Schloter and C. M. Thomas, “Increased Abundance of IncP-1β Plasmids and Mercury Resistance Genes in Mercury-Polluted River Sediments: First Discovery of IncP-1β Plasmids with a Complex mer Transposon as the Sole Accessory Element,” Applied and En vironmental Microbiology, Vol. 72, No. 11, 2006, pp. 7253-7259. doi:10.1128/AEM.00922-06
- [89] M. S. Wright, C. Baker-Austin, A. H. Lindell, R. Stepanauskas, H. W. Stokes and J. V. McArthur, “Influence of Industrial Contamination on Mobile Genetic Elements: Class 1 Integron Abundance and Gene Cassette Structure in Aquatic Bacterial Communities,” The ISME Journal, Vol. 2, No. 4, 2008, pp. 417-428. doi:10.1038/ismej.2008.8
- [90] C. P. Rosewarne, V. Pettigrove, H. W. Stokes and Y. M. Parsons, “Class 1 Integrons in Benthic Bacterial Communities: Abundance, Association with Tn402-Like Transposition Modules and Evidence for Coselection with Heavy-Metal Resistance,” FEMS Microbiology Ecology, Vol. 72, No. 1, 2010, pp. 35-46. doi:10.1111/j.1574-6941.2009.00823.x
- [91] N. F. Burton, M. J. Day and A. T. Bull, “Distribution of Bacterial Plasmids in Clean and Polluted Sites in a South Wales River,” Applied and Environmental Microbiology, Vol. 44, No. 5, 1982, pp. 1026-1029.
- [92] R. P. Schwarzenbach, B. I. Escher, K. Fenner, T. B. Hofstetter, C. A. Johnson, U. von Gunten and B. Wehrli, “The Challenge of Micropollutants in Aquatic Systems,” Science, Vol. 313, No. 5790, 2006, pp. 1072-1077. doi:10.1126/science.1127291
- [93] K. Kümmerer, “Antibiotics in the Aquatic Environment—A Review—Part I,” Chemosphere, Vol. 75, No. 4, 2009, pp. 417-434. doi:10.1016/j.chemosphere.2008.11.086
- [94] B. L. Skjelkvåle, T. Andersen, E. Fjeld, J. Mannio, A. Wilander, K. Johansson, J. P. Jensen and T. Moiseenko, “Heavy Metal Surveys in Nordic Lakes; Concentrations, Geographic Patterns and Relation to Critical Limits,” Ambio, Vol. 30, No. 1, 2001, pp. 2-10.
- [95] A. C. Alder, A. Bruchet, M. Carballa, M. Clara, A. Joss, D. Löffler, C. S. McArdell, K. Miksch, F. Omil, T. Tuhkanen and T. A. Ternes, “Consumption and Occurrence,” In: T. A. Ternes and A. Joss, Eds., Human Pharmaceuticals, Hormones and Fragrances: The Challenge of Micropollutants in Urban Water Management, IWA Publishing, London, 2009, pp. 15-54.
- [96] D. E. Armstrong and A. W. Elzerman, “Trace Metal Accumulation in Surface Microlayers,” Journal of Great Lakes Research, Vol. 8, No. 2, 1982, pp. 282-287. doi:10.1016/S0380-1330(82)71966-5
- [97] J. Eggleton and K. V. Thomas, “A Review of Factors Affecting the Release and Bioavailability of Contaminants during Sediment Disturbance Events,” Environment International, Vol. 30, No. 7, 2004, pp. 973-980. doi:10.1016/j.envint.2004.03.001
- [98] O. Wurl and J. P. Obbard, “A Review of Pollutants in the Sea-Surface Microlayer (SML): A Unique Habitat for Marine Organisms,” Marine Pollution Bulletin, Vol. 48, No. 11-12, 2004, pp. 1016-1030. doi:10.1016/j.marpolbul.2004.03.016
- [99] E. Haack and L. A. Warren, “Biofilm Hydrous Manganese Oxyhydroxides and Metal Dynamics in Acid Rock Drainage,” Environmental Science and Technology, Vol. 37, No. 18, 2003, pp. 4138-4147. doi:10.1021/es026274z
- [100] D. B. Wunder, V. A. Bosscher, R. C. Cok and R. M. Hozalski, “Sorption of Antibiotics to Biofilm,” Water Research, Vol. 45, No. 6, 2011, pp. 2270-2280. doi:10.1016/j.watres.2010.11.013
- [101] J. M. Harrington, S. E. Fendorf and R. F. Rosenzweig, “Biotic Generation of Arsenic(III) in Metal(loid)-Contaminated Freshwater Lake Sediments,” Environmental Science and Technology, Vol. 32, No. 16, 1998, pp. 2425- 2430. doi:10.1021/es971129k
- [102] R. P. Rastogi, Richa, A. Kumar, M. B. Tyagi and R. P. Sinha, “Molecular Mechanisms of Ultraviolet RadiationInduced DNA Damage and Repair,” Journal of Nucleic Acids, 2010, Article ID: 592980. doi:10.4061/2010/592980
- [103] R. Maranger, P. A. del Giorgio and D. F. Bird, “Accumulation of Damaged Bacteria and Viruses in Lake Water Exposed to Solar Radiation,” Aquatic Microbial Ecology, Vol. 28, No. 3, 2002, pp. 213-227. doi:10.3354/ame028213
- [104] K. E. Wommack, R. T. Hill, T. A. Muller and R. R. Colwell, “Effects of Sunlight on Bacteriophage Viability and Structure,” Applied and Environmental Microbiology, Vol. 62, No. 4, 1996, pp. 1336-1341.
- [105] S. W. Wilhelm, M. G. Weinbauer, C. A. Suttle and W. H. Jeffrey, “The Role of Sunlight in the Removal and Repair of Viruses in the Sea,” Limnology and Oceanography, Vol. 43, No. 4, 1998, pp. 586-592. doi:10.4319/lo.1998.43.4.0586
- [106] J. L. Ravanat, T. Douki and J. Cadet, “Direct and Indirect Effects of UV Radiation on DNA and Its Components,” Journal of Photochemistry and Photobiology B: Biology, Vol. 63, No. 1-3, 2001, pp. 88-102. doi:10.1016/S1011-1344(01)00206-8
- [107] M. A. Tapper and R. E. Hicks, “Temperate Viruses and Lysogeny in Lake Superior Bacterioplankton,” Limnology and Oceanography, Vol. 43, No. 1, 1998, pp. 95-103. doi:10.4319/lo.1998.43.1.0095
- [108] S. W. Wilhelm and R. E. H. Smith, “Bacterial Carbon Production in Lake Erie is Influenced by Viruses and Solar Radiation,” Canadian Journal of Fisheries and Aquatic Sciences, Vol. 57, No. 2, 2000, pp. 317-326. doi:10.1139/f99-202
- [109] D. P. Häder, H. D. Kumar, R. C. Smith and R. C. Worrest, “Effects of Solar UV Radiation on Aquatic Ecosystems and Interactions with Climate Change,” Photochemical and Photobiological Sciences, Vol. 6, No. 3, 2007, pp. 267-285. doi:10.1039/b700020k
- [110] D. P. Morris, H. Zagarese, C. E. Williamson, E. G. Balseiro, B. R. Hargreaves, B. Modenutti, R. Moeller and C. Queimalinos, “The Attenuation of Solar UV Radiation in Lakes and the Role of Dissolved Organic Carbon,” Limnology and Oceanography, Vol. 40, No. 8, 1995, pp. 1381-1391. doi:10.4319/lo.1995.40.8.1381
- [111] N. M. Scully and D. R. S. Lean, “The Attenuation of Ultraviolet Radiation in Temperate Lakes,” Archiv für Hydrobiologie, Vol. 43, 1994, pp. 135-144.
- [112] M. G. Lorenz, B. W. Aardema and W. Wackernagel, “Highly Efficient Genetic Transformation of Bacillus subtilis Attached to Sand Grains,” Journal of General Microbiology, Vol. 134, No. 1, 1988, pp. 107-112.
- [113] B. Chamier, M. G. Lorenz and W. Wackernagel, “Natural Transformation of Acinetobacter calcoaceticus by Plasmid DNA Adsorbed on Sand and Groundwater Aquifer Material,” Applied and Environmental Microbiology, Vol. 59, No. 5, 1993, pp. 1662-1667.
- [114] C. Vettori, G. Stotzky, M. Yoder and E. Gallori, “Interaction between Bacteriophage PBS1 and Clay Minerals and Transduction of Bacillus subtilis by Clay-Phage Complexes,” Environmental Microbiology, Vol. 1, No. 4, 1999, pp. 347-355. doi:10.1046/j.1462-2920.1999.00044.x
- [115] C. Vettori, E. Gallori and G. Stotzky, “Clay Minerals Protect Bacteriophage PBS1 of Bacillus subtilis Against Inactivation and Loss of Transducing Ability by UV Radiation,” Canadian Journal of Microbiology, Vol. 46, No. 8, 2000, pp. 770-773.
- [116] D. Kirchman and R. Mitchell, “Contribution of Particle-Bound Bacteria to Total Microheterotrophic Activity in Five Ponds and Two Marshes,” Applied and Environmental Microbiology, Vol. 43, No. 1, 1982, pp. 200-209.
- [117] B. C. Crump, J. A. Baross and C. A. Simenstad, “Dominance of Particle-Attached Bacteria in the Columbia River Estuary, USA,” Aquatic Microbial Ecology, Vol. 14, No. 1, 1998, pp. 7-18. doi:10.3354/ame014007
- [118] N. D. Seeley and S. B. Primrose, “A Review: The Isolation of Bacteriophages from the Environment,” Journal of Applied Bacteriology, Vol. 53, No. 1, 1982, pp. 1-17. doi:10.1111/j.1365-2672.1982.tb04729.x
- [119] L. Kernegger, I. Zweimüller and P. Peduzzi, “Effects of Suspended Matter Quality and Virus Abundance on Microbial Parameters: Experimental Evidence from a Large European River,” Aquatic Microbial Ecology, Vol. 57, No. 2, 2009, pp. 161-173. doi:10.3354/ame01341
- [120] G. Pietramellara, J. Ascher, F. Borgogni, M. T. Ceccherini, G. Guerri and P. Nannipieri, “Extracellular DNA in Soil and Sediment: Fate and Ecological Relevance,” Biology and Fertility of Soils, Vol. 45, No. 3, 2009, pp. 219-235. doi:10.1007/s00374-008-0345-8
- [121] G. Romanowski, M. G. Lorenz and W. Wackernagel, “Plasmid DNA in a Groundwater Aquifer Microcosm— Adsorption, DNAase Resistance and Natural Genetic Transformation of Bacillus subtilis,” Molecular Ecology, Vol. 2, No. 3, 1993, pp. 171-181. doi:10.1111/j.1365-294X.1993.tb00106.x
- [122] C. Crecchio and G. Stotzky, “Binding of DNA on Humic Acids: Effect on Transformation of Bacillus subtilis and Resistance to DNase,” Soil Biology and Biochemistry, Vol. 30, No. 8-9, 1998, pp. 1061-1067. doi:10.1016/S0038-0717(97)00248-4
- [123] N. Lu, J. L. Zilles and T. H. Nguyen, “Adsorption of Extracellular Chromosomal DNA and Its Effects on Natural Transformation of Azotobacter vinelandii,” Applied and Environmental Microbiology, Vol. 76, No. 13, 2010, pp. 4179-4184. doi:10.1128/AEM.00193-10
- [124] P. Cai, Q. Huang, W. Chen, D. Zhang, K. Wang, D. Jiang and W. Liang, “Soil Colloids-Bound Plasmid DNA: Effect on Transformation of E. coli and Resistance to DNase I Degradation,” Soil Biology and Biochemistry, Vol. 39, No. 5, 2007, pp. 1007-1013. doi:10.1016/j.soilbio.2006.11.010
- [125] K. E. Hill and E. M. Top, “Gene Transfer in Soil Systems Using Microcosms,” FEMS Microbiology Ecology, Vol. 25, No. 4, 1998, pp. 319-329. doi:10.1111/j.1574-6941.1998.tb00483.x
- [126] J. M. Plach, A. V. C. Elliott, I. G. Droppo and L. A. Warren, “Physical and Ecological Controls on Freshwater Floc Trace Metal Dynamics,” Environmental Science and Technology, Vol. 45, No. 6, 2011, pp. 2157-2164. doi:10.1021/es1031745
- [127] A. V. C. Elliott, J. M. Plach, I. G. Droppo and L. A. Warren, “Comparative Floc-Bed Sediment Trace Element Partitioning across Variably Contaminated Aquatic Ecosystems,” Environmental Science and Technology, Vol. 46, No. 1, 2012, pp. 209-216. doi:10.1021/es202221u
- [128] Z. Qiu, Y. Yu, Z. Chen, M. Jin, D. Yang, Z. Zhao, J. Wang, Z. Shen, X. Wang, D. Qian, A. Huang, B. Zhang and J. W. Li, “Nanoalumina Promotes the Horizontal Transfer of Multiresistance Genes Mediated by Plasmids across Genera,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 109, No. 13, 2012, pp. 4944-4949. doi:10.1073/pnas.1107254109
- [129] A. Södergren, “Role of Aquatic Surface Microlayer in the Dynamics of Nutrients and Organic Compounds in Lakes, with Implications for the Ecotones,” Hydrobiologia, Vol. 251, No. 1-3, 1993, pp. 217-225. doi:10.1007/BF00007181
- [130] G. A. Weyhenmeyer, L. Hakanson and M. Meili, “A Validated Model for Daily Variations in the Flux, Origin, and Distribution of Settling Particles Within Lakes,” Limnology and Oceanography, Vol. 42, No. 7, 1997, pp. 1517-1529. doi:10.4319/lo.1997.42.7.1517
- [131] M. K. Amin and M. J. Day, “Influence of pH Value on Viability and Transduction Frequency of Pseudomonas aeruginosa Phage F116,” Letters in Applied Microbiology, Vol. 6, No. 4, 1988b, pp. 93-96. doi:10.1111/j.1472-765X.1988.tb01222.x
- [132] P. A. Rochelle, M. J. Day and J. C. Fry, “Occurrence, Transfer and Mobilization in Epilithic Strains of Acinetobacter of Mercury-Resistance Plasmids Capable of Transformation,” Journal of General Microbiology, Vol. 134, No. 11, 1988, pp. 2933-2941.
- [133] P. A. Rochelle, J. C. Fry and M. J. Day, “Factors Affecting Conjugal Transfer of Plasmids Encoding Mercury Resistance from Pure Cultures and Mixed Natural Suspensions of Epilithic Bacteria,” Journal of General Microbiology, Vol. 135, No. 2, 1989, pp. 409-424.
- [134] A. Fernandez-Astorga, A. Muela, R. Cisterna, J. Iriberri and I. Barcina, “Biotic and Abiotic Factors Affecting Plasmid Transfer in Escherichia coli Strains,” Applied and Environmental Microbiology, Vol. 58, No. 1, 1992, pp. 392-398.
- [135] L. J. Tranvik, W. Graneli and G. Gahnstroem, “Microbial Activity in Acidified and Limed Humic Lakes,” Canadian Journal of Fisheries and Aquatic Sciences, Vol. 51, No. 11, 1994, pp. 2529-2536. doi:10.1139/f94-252
- [136] S. C. Maberly, “Diel, Episodic and Seasonal Changes in pH and Concentrations of Inorganic Carbon in a Productive Lake,” Freshwater Biology, Vol. 35, No. 3, 1996, pp. 579-598. doi:10.1111/j.1365-2427.1996.tb01770.x
- [137] A. L. Mills, J. S. Herman, G. M. Hornberger and T. H. DeJesús, “Effect of Solution Ionic Strength and Iron Coatings on Mineral Grains on the Sorption of Bacterial Cells to Quartz Sand,” Applied and Environmental Microbiology, Vol. 60, No. 9, 1994, pp. 3300-3306.
- [138] T. H. Nguyen and K. L. Chen, “Role of Divalent Cations in Plasmid DNA Adsorption to Natural Organic Matter-Coated Silica Surface,” Environmental Science and Technology, Vol. 41, No. 15, 2007, pp. 5370-5375. doi:10.1021/es070425m
- [139] X. Wang, M. Li, Q. Yan, X. Chen, J. Geng, Z. Xie and P. Shen, “Across Genus Plasmid Transformation between Bacillus subtilis and Escherichia coli and the Effect of Escherichia coli on the Transforming Ability of Free Plasmid DNA,” Current Microbiology, Vol. 54, No. 6, 2007, pp. 450-456. doi:10.1007/s00284-006-0617-1
- [140] W. J. Page and M. von Tigerstrom, “Optimal Conditions for Transformation of Azotobacter vinelandii,” Journal of Bacteriology, Vol. 139, No. 3, 1979, pp. 1058-1061.
- [141] B. Baur, K. Hanselmann, W. Schlimme and B. Jenni, “Genetic Transformation in Freshwater: Escherichia coli Is Able to Develop Natural Competence,” Applied and Environmental Microbiology, Vol. 62, No. 10, 1996, pp. 3673-3678.
- [142] J. M. Solomon and A. D. Grossman, “Who’s Competent and When: Regulation of Natural Genetic Competence in Bacteria,” Trends in Genetics, Vol. 12, No. 4, 1996, pp. 150-155. doi:10.1016/0168-9525(96)10014-7
- [143] M. C. Trombe, C. Clavé and J. M. Manias, “Calcium Regulation of Growth and Differentiation in Streptococcus pneumonia,” Journal of General Microbiology, Vol. 138, No. 1, 1992, pp. 77-84. doi:10.1099/00221287-138-1-77
- [144] C. M. Thomas and K. M. Nielsen, “Mechanisms of, and Barriers to, Horizontal Gene Transfer between Bacteria,” Nature Reviews Microbiology, Vol. 3, No. 9, 2005, pp. 711-721. doi:10.1038/nrmicro1234
- [145] J. Replicon, A. Frankfater and R. V. Miller, “A Continuous Culture Model to Examine Factors that Affect Transduction among Pseudomonas aeruginosa Strains in Freshwater Environments,” Applied and Environmental Microbiology, Vol. 61, No. 9, 1995, pp. 3359-3366.
- [146] P. Tuomi, T. Torsvik, M. Heldal and G. Bratbak, “Bacterial Population Dynamics in a Meromictic Lake,” Applied and Environmental Microbiology, Vol. 63, No. 6, 1997, pp. 2181-2188.
- [147] M. G. Weinbauer and M. G. Höfle, “Significance of Viral Lysis and Flagellate Grazing as Factors Controlling Bacterioplankton Production in a Eutrophic Lake,” Applied and Environmental Microbiology, Vol. 64, No. 2, 1998, pp. 431-438.
- [148] M. G. Weinbauer and M. G. Höfle, “Size-Specific Mortality of Lake Bacterioplankton by Natural Virus Communities,” Aquatic Microbial Ecology, Vol. 15, No. 2, 1998, pp. 103-113. doi:10.3354/ame015103
- [149] Y. Bettarel, T. Sime-Ngando, C. Amblard and J. Dolan, “Viral Activity in Two Contrasting Lake Ecosystems,” Applied and Environmental Microbiology, Vol. 70, No. 5, 2004, pp. 2941-2951. doi:10.1128/AEM.70.5.2941-2951.2004
- [150] J. W. Costerton, Z. Lewandowski, D. E. Caldwell, D. R. Korber and H. M. Lappin-Scott, “Microbial Biofilms,” Annual Review of Microbiology, Vol. 49, 1995, pp. 711- 745. doi:10.1146/annurev.mi.49.100195.003431
- [151] S. J. Sørensen, M. Bailey, L. H. Hansen, N. Kroer and S. Wuertz, “Studying Plasmid Horizontal Transfer in Situ: A Critical Review,” Nature Reviews Microbiology, Vol. 3, No. 9, 2005, pp. 700-710. doi:10.1038/nrmicro1232
- [152] K. B. Barken, S. J. Pamp, L. Yang, M. Gjermansen, J. J. Bertrand, M. Klausen, M. Givskov, C. B. Whitchurch, J. N. Engel and T. Tolker-Nielsen, “Roles of Type IV Pili, Flagellum-Mediated Motility and Extracellular DNA in the Formation of Mature Multicellular Structures in Pseudomonas aeruginosa Biofilms,” Environmental Microbiology, Vol. 10, No. 9, 2008, pp. 2331-2343. doi:10.1111/j.1462-2920.2008.01658.x
- [153] J. M. Ghigo, “Natural Conjugative Plasmids Induce Bacterial Biofilm Development,” Nature, Vol. 412, No. 6845, 2001, pp. 442-445. doi:10.1038/35086581
- [154] J. Gödeke, K. Paul, J. Lassak and K. M. Thormann, “Phage-Induced Lysis Enhances Biofilm Formation in Shewanella oneidensis MR-1,” The ISME Journal, Vol. 5, No. 4, 2011, pp. 613-626. doi:10.1038/ismej.2010.153
- [155] M. Hausner and S. Wuertz, “High Rates of Conjugation in Bacterial Biofilms as Determined by Quantitative in Situ Analysis,” Applied and Environmental Microbiology, Vol. 65, No. 8, 1999, pp. 3710-3713.
- [156] S. Molin and T. Tolker-Nielsen, “Gene Transfer Occurs with Enhanced Efficiency in Biofilms and Induces Enhanced Stabilisation of the Biofilm Structure,” Current Opinion in Biotechnology, Vol. 14, No. 3, 2003, pp. 255- 261. doi:10.1016/S0958-1669(03)00036-3
- [157] S. Maeda, M. Ito, T. Ando, Y. Ishimoto, Y. Fujisawa, H. Takahashi, A. Matsuda, A. Sawamura and S. Kato, “Horizontal Transfer of Nonconjugative Plasmids in a Colony Biofilm of Escherichia coli,” FEMS Microbiology Letters, Vol. 255, No. 1, 2006, pp. 115-120. doi:10.1111/j.1574-6968.2005.00072.x
- [158] R. E. Fox, X. Zhong, S. M. Krone and E. M. Top, “Spatial Structure and Nutrients Promote Invasion of IncP-1 Plasmids in Bacterial Populations,” The ISME Journal, Vol. 2, No. 10, 2008, pp. 1024-1039. doi:10.1038/ismej.2008.53
- [159] M. L. Angles, K. C. Marshall and A. E. Goodman, “Plasmid Transfer between Marine Bacteria in the Aqueous Phase and Biofilms in Reactor Microcosms,” Applied and Environmental Microbiology, Vol. 59, No. 3, 1993, pp. 843-850.
- [160] M. K. Kay, T. C. Erwin, R. J. C. McLean and G. M. Aron, “Bacteriophage Ecology in Escherichia coli and Pseudomonas aeruginosa Mixed-Biofilm Communities,” Applied and Environmental Microbiology, Vol. 77, No. 3, 2011, pp. 821-829. doi:10.1128/AEM.01797-10
- [161] A. Resch, B. Fehrenbacher, K. Eisele, M. Schaller and F. Götz, “Phage Release from Biofilm and Planktonic Staphylococcus aureus Cells,” FEMS Microbiology Letters, Vol. 252, No. 1, 2005, pp. 89-96. doi:10.1016/j.femsle.2005.08.048
- [162] P. S. Stewart and M. J. Franklin, “Physiological Heterogeneity in Biofilms,” Nature Reviews Microbiology, Vol. 6, No. 3, 2008, pp. 199-210. doi:10.1038/nrmicro1838
- [163] G. Hidalgo, A. Burns, E. Herz, A. G. Hay, P. L. Houston, U. Wiesner and L. W. Lion, “Functional Tomographic Fluorescence Imaging of pH Microenvironments in Microbial Biofilms by Use of Silica Nanoparticle Sensors,” Applied and Environmental Microbiology, Vol. 75, No. 23, 2009, pp. 7426-7435. doi:10.1128/AEM.01220-09
- [164] A. P. Hitchcock, J. J. Dynes, J. R. Lawrence, M. Obst, G. D. W. Swerhone, D. R. Korber and G. G. Leppard, “Soft X-Ray Spectromicroscopy of Nickel Sorption in a Natural River Biofilm,” Geobiology, Vol. 7, No. 4, 2009, 432-453. doi:10.1111/j.1472-4669.2009.00211.x
- [165] G. Chen, X. Chen, Y. Yang, A. G. Hay, X. Yu and Y. Chen, “Sorption and Distribution of Copper in Unsaturated Pseudomonas putida CZ1 Biofilms as Determined by X-ray Fluorescence Microscopy,” Applied and Environmental Microbiology, Vol. 77, No. 14, 2011, pp. 4719- 4727. doi:10.1128/AEM.00125-11
- [166] J. D. van Elsas and M. J. Bailey, “The Ecology of Transfer of Mobile Genetic Elements,” FEMS Microbiology Ecology, Vol. 42, No. 2, 2002, pp. 187-197.
- [167] M. Hermansson and C. Linberg, “Gene Transfer in the Marine Environment,” FEMS Microbiology Ecology, Vol. 15, No. 1-2, 1994, pp. 47-54. doi:10.1111/j.1574-6941.1994.tb00228.x
- [168] B. F. Smets, B. E. Rittmann and D. A. Stahl, “Quantification of the Effect of Substrate Concentration on the Conjugal Transfer Rate of the TOL Plasmid in Short-Term Batch Mating Experiments,” Letters in Applied Microbiology, Vol. 21, No. 3, 1995, pp. 167-172. doi:10.1111/j.1472-765X.1995.tb01033.x
- [169] M. R. Shakibaie, K. A. Jalilzadeh and S. M. Yamakanamardi, “Horizontal Transfer of Antibiotic Resistance Genes among Gram Negative Bacteria in Sewage and Lake Water and Influence of Some Physico-Chemical Parameters of Water on Conjugation Process,” Journal of Environmental Biology, Vol. 30, No. 1, 2009, pp. 45-49.
- [170] B. Normander, B. B. Christensen, S. Molin and N. Kroer, “Effect of Bacterial Distribution and Activity on Conjugal Gene Transfer on the Phylloplane of the Bush Bean (Phaseolus vulgaris),” Applied and Environmental Microbiology, Vol. 64, No. 5, 1998, pp. 1902-1909.
- [171] A. R. Johnsen and N. Kroer, “Effects of Stress and Other Environmental Factors on Horizontal Plasmid Transfer Assessed by Direct Quantification of Discrete Transfer Events,” FEMS Microbiology Ecology, Vol. 59, No. 3, 2007, pp. 718-728. doi:10.1111/j.1574-6941.2006.00230.x
- [172] T. A. Kokjohn and G. S. Sayler, “Attachment and Replication of Pseudomonas aeruginosa Bacteriophages under Conditions Simulating Aquatic Environments,” Journal of General Microbiology, Vol. 137, No. 3, 1991, pp. 661- 666. doi:10.1099/00221287-137-3-661
- [173] S. E. Finkel and R. Kolter, “DNA as a Nutrient: Novel Role for Bacterial Competence Gene Homologs,” Journal of Bacteriology, Vol. 183, No. 21, 2001, pp. 6288-6293. doi:10.1128/JB.183.21.6288-6293.2001
- [174] J. J. Elser, L. B. Stabler and R. P. Hassett, “Nutrient Limitation of Bacterial Growth and Rates of Bacterivory in Lakes and Oceans: A Comparative Study,” Aquatic Microbial Ecology, Vol. 9, No. 2, 1995, pp. 105-110. doi:10.3354/ame009105
- [175] E. M. Smith and Y. T. Prairie, “Bacterial Metabolism and Growth Efficiency in Lakes: The Importance of Phosphorus Availability,” Limnology and Oceanography, Vol. 49, No. 1, 2004, pp. 137-147. doi:10.4319/lo.2004.49.1.0137
- [176] N. E. Melechen and G. Go, “Induction of Lambdoid Prophages by Amino Acid Deprivation: Differential Inducibility; Role of recA,” Molecular and General Genetics, Vol. 180, No. 1, 1980, pp. 147-155. doi:10.1007/BF00267364
- [177] J. H. Paul, M. E. Frischer and J. M. Thurmond, “Gene Transfer in Marine Water Column and Sediment Microcosms by Natural Plasmid Transformation,” Applied and Environmental Microbiology, Vol. 57, No. 5, 1991a, pp. 1509-1515.
- [178] A. E. Goodman, K. C. Marshall and M. Hermansson, “Gene Transfer among Bacteria under Conditions of Nutrient Depletion in Simulated and Natural Aquatic Environments,” FEMS Microbiology Ecology, Vol. 15, No. 1- 2, 1994, pp. 55-60. doi:10.1111/j.1574-6941.1994.tb00229.x
- [179] J. J. Cole, M. L. Pace, N. F. Caraco and G. S. Steinhart, “Bacterial Biomass and Cell Size Distributions in Lakes: More and Larger Cells in Anoxic Waters,” Limnology and Oceanography, Vol. 38, No. 8, 1993, pp. 1627-1632. doi:10.4319/lo.1993.38.8.1627
- [180] P. Carlsson and D. A. Caron, “Seasonal Variation of Phosphorus Limitation of Bacterial Growth in a Small Lake,” Limnology and Oceanography, Vol. 46, No. 1, 2001, pp. 108-120. doi:10.4319/lo.2001.46.1.0108
- [181] U. Münster, E. Heikkinen and J. Knulst, “Nutrient Composition, Microbial Biomass and Activity at the Air-Water Interface of Small Boreal Forest Lakes,” Hydrobiologia, Vol. 363, No. 1-3, 1997, pp. 261-270.
- [182] B. Boström, J. M. Andersen, S. Fleischer and M. Jansson, “Exchange of Phosphorus across the Sediment-Water Interface,” Hydrobiologia, Vol. 170, No. 1, 1988, pp. 229- 244. doi:10.1007/BF00024907
- [183] J. E. González and M. M. Marketon, “Quorum Sensing in Nitrogen-Fixing Rhizobia,” Microbiology and Molecular Biology Reviews, Vol. 67, No. 4, 2003, pp. 574-592. doi:10.1128/MMBR.67.4.574-592.2003
- [184] J. P. Ramsay, J. T. Sullivan, N. Jambari, C. A. Ortori, S. Heeb, P. Williams, D. A. Barrett, I. L. Lamont and C. W. Ronson, “A LuxRI-Family Regulatory System Controls Excision and Transfer of the Mesorhizobium loti Strain R7A Symbiosis Island by Activating Expression of Two Conserved Hypothetical Genes,” Molecular Microbiology, Vol. 73, No. 6, 2009, pp. 1141-1155. doi:10.1111/j.1365-2958.2009.06843.x
- [185] D. Ghosh, K. Roy, K. E. Williamson, S. Srinivasiah, K. E. Wommack and M. Radosevich, “Acyl-Homoserine Lactones Can Induce Virus Production in Lysogenic Bacteria: An Alternative Paradigm for Prophage Induction,” Applied and Environmental Microbiology, Vol. 75, No. 22, 2009, pp. 7142-7152. doi:10.1128/AEM.00950-09
- [186] E. S. Antonova and B. K. Hammer, “Quorum-Sensing Autoinducer Molecules Produced by Members of a Multispecies Biofilm Promote Horizontal Gene Transfer to Vibrio cholerae,” FEMS Microbiology Letters, Vol. 322, No. 1, 2011, pp. 68-76. doi:10.1111/j.1574-6968.2011.02328.x
- [187] L. Mølbak, S. Molin and N. Kroer, “Root Growth and Exudate Production Define the Frequency of Horizontal Plasmid Transfer in the Rhizosphere,” FEMS Microbiology Ecology, Vol. 59, No. 1, 2007, pp. 167-176. doi:10.1111/j.1574-6941.2006.00229.x
- [188] N. Kroer, T. Barkay, S. Sørensen and D. Weber, “Effect of Root Exudates and Bacterial Metabolic Activity on Conjugal Gene Transfer in the Rhizosphere of a Marsh Plant,” FEMS Microbiology Ecology, Vol. 25, No. 4, 1998, pp. 375-384. doi:10.1111/j.1574-6941.1998.tb00489.x
- [189] M. Ueki, K. Matsui, K. Choi and Z. Kawabata, “The Enhancement of Conjugal Plasmid pBHR1 Transfer between Bacteria in the Presence of Extracellular Metabolic Products Produced by Microcystis aeruginosa,” FEMS Microbiology Ecology, Vol. 51, No. 1, 2004, pp. 1-8. doi:10.1016/j.femsec.2004.07.003
- [190] K. Matsui, N. Ishii and Z. Kawabata, “Release of Extracellular Transformable Plasmid DNA from Escherichia coli Cocultivated with Algae,” Applied and Environmental Microbiology, Vol. 69, No. 4, 2003, pp. 2399- 2404. doi:10.1128/AEM.69.4.2399-2404.2003
- [191] K. Matsui, N. Ishii and Z. Kawabata, “Microbial Interactions Affecting the Natural Transformation of Bacillus subtilis in a Model Aquatic Ecosystem,” FEMS Microbiology Ecology, Vol. 45, No. 3, 2003, pp. 211-218. doi:10.1016/S0168-6496(03)00148-X
- [192] J. Matsuo, S. Oguri, S. Nakamura, T. Hanawa, T. Fukumoto, Y. Hayashi, K. Kawaguchi, Y. Mizutani, T. Yao, K. Akizawa, H. Suzuki, C. Simizu, K. Matsuno, S. Kamiya and H. Yamaguchi, “Ciliates Rapidly Enhance the Frequency of Conjugation between Escherichia coli Strains through Bacterial Accumulation in Vesicles,” Research in Microbiology, Vol. 161, No. 8, 2010, pp. 711-719. doi:10.1016/j.resmic.2010.07.004
- [193] T. R. de Kievit, “Quorum Sensing in Pseudomonas aeruginosa Biofilms,” Environmental Microbiology, Vol. 11, No. 2, 2009, pp. 279-288. doi:10.1111/j.1462-2920.2008.01792.x
- [194] P. Cai, Q. Huang, J. Zhu, D. Jiang, X. Zhou, X. Rong and W. Liang, “Effects of Low-Molecular-Weight Organic Ligands and Phosphate on DNA Adsorption by Soil Colloids and Minerals,” Colloids and Surfaces B: Biointerfaces, Vol. 54, No. 1, 2007, pp. 53-59. doi:10.1016/j.colsurfb.2006.07.013
- [195] K. L. Meibom, M. Blokesch, N. A. Dolganov, C. Y. Wu and G. K. Schoolnik, “Chitin Induces Natural Competence in Vibrio cholerae,” Science, Vol. 310, No. 5755, 2005, pp. 1824-1827. doi:10.1126/science.1120096
- [196] H. W. Paerl and J. L. Pinckney, “A Mini-Review of Microbial Consortia: Their Roles in Aquatic Production and Biogeochemical Cycling,” Microbial Ecology, Vol. 31, No. 3, 1996, pp. 225-247. doi:10.1007/BF00171569
- [197] L. de Gelder, F. P. Vandecasteele, C. J. Brown, L. J. Forney and E. M. Top, “Plasmid Donor Affects Host Range of Promiscuous IncP-1β Plasmid pB10 in an ActivatedSludge Microbial Community,” Applied and Environmental Microbiology, Vol. 71, No. 9, 2005, pp. 5309- 5317. doi:10.1128/AEM.71.9.5309-5317.2005
- [198] I. Chen and D. Dubnau, “DNA Uptake during Bacterial Transformation,” Nature Reviews Microbiology, Vol. 2, No. 3, 2004, pp. 241-249. doi:10.1038/nrmicro844
- [199] A. C. Yannarell and E. W. Triplett, “Withinand Between-Lake Variability in the Composition of Bacterioplankton Communities: Investigations Using Multiple Spatial Scales,” Applied and Environmental Microbiology, Vol. 70, No. 1, 2004, pp. 214-223. doi:10.1128/AEM.70.1.214-223.2004
- [200] R. C. Crump, H. E. Adams, J. E. Hobbie and G. W. Kling, “Biogeography of Bacterioplankton in Lakes and Streams of an Arctic Tundra Catchment,” Ecology, Vol. 88, No. 6, 2007, pp. 1365-1378. doi:10.1890/06-0387
- [201] K. Dominik and M. G. Höfle, “Changes in Bacterioplankton Community Structure and Activity with Depth in a Eutrophic Lake as Revealed by 5S rRNA Analysis,” Applied and Environmental Microbiology, Vol. 68, No. 7, 2002, pp. 3606-3613. doi:10.1128/AEM.68.7.3606-3613.2002
- [202] A. C. Yannarell, A. D. Kent, G. H. Lauster, T. K. Kratz and E. W. Triplett, “Temporal Patterns in Bacterial Communities in Three Temperate Lakes of Different Trophic Status,” Microbial Ecology, Vol. 46, No. 4, 2003, pp. 391-405. doi:10.1007/s00248-003-1008-9
- [203] W. Zwisler, N. Selje and M. Simon, “Seasonal Patterns of the Bacterioplankton Community Composition in a Large Mesotrophic Lake,” Aquatic Microbial Ecology, Vol. 31, No. 3, 2003, pp. 211-225. doi:10.3354/ame031211
- [204] S. R. Mueller-Spitz, G. W. Goetz and S. L. McLellan, “Temporal and Spatial Variability in Nearshore Bacterioplankton Communities of Lake Michigan,” FEMS Microbiology Ecology, Vol. 67, No. 3, 2009, pp. 511-522. doi:10.1111/j.1574-6941.2008.00639.x
- [205] A. Konopka, T. Bercot and C. Nakatsu, “Bacterioplankton Community Diversity in a Series of Thermally Stratified Lakes,” Microbial Ecology, Vol. 38, No. 2, 1999, pp. 126-135. doi:10.1007/s002489900166
- [206] D. Boucher, L. Jardillier and D. Debroas, “Succession of Bacterial Community Composition over Two Consecutive Years in Two Aquatic Systems: A Natural Lake and a Lake-Reservoir,” FEMS Microbiology Ecology, Vol. 55, No. 1, 2006, pp. 79-97. doi:10.1111/j.1574-6941.2005.00011.x
- [207] A. Shade, S. E. Jones and K. D. McMahon, “The Influence of Habitat Heterogeneity on Freshwater Bacterial Community Composition and Dynamics,” Environmental Microbiology, Vol. 10, No. 4, 2008, pp. 1057-1067. doi:10.1111/j.1462-2920.2007.01527.x
- [208] E. F. DeLong, D. G. Franks and A. L. Alldredge, “Phylogenetic Diversity of Aggregate-Attached vs. Free-Living Marine Bacterial Assemblages,” Limnology and Oceanography, Vol. 38, No. 5, 1993, pp. 924-934. doi:10.4319/lo.1993.38.5.0924
- [209] B. C. Crump, E. V. Armbrust and J. A. Baross, “Phylogenetic Analysis of Particle-Attached and Free-Living Bacterial Communities in the Columbia River, Its Estuary, and the Adjacent Coastal Ocean,” Applied and Environmental Microbiology, Vol. 65, No. 7, 1999, pp. 3192- 3204.
- [210] D. A. Pearce, C. J. van der Gast, K. Woodward and K. K. Newsham, “Significant Changes in the Bacterioplankton Community Structure of a Maritime Antarctic Freshwater Lake Following Nutrient Enrichment,” Microbiology, Vol. 151, No. 10, 2005, pp. 3237-3248. doi:10.1099/mic.0.27258-0
- [211] J. A. J. Haagensen, S. K. Hansen, T. Johansen and S. Molin, “In Situ Detection of Horizontal Transfer of Mobile Genetic Elements,” FEMS Microbiology Ecology, Vol. 42, No. 2, 2002, pp. 261-268. doi:10.1111/j.1574-6941.2002.tb01016.x
- [212] M. C. Haug, S. A. Tanner, C. Lacroix, L. Meile and M. J. A. Stevens, “Construction and Characterization of Enterococcus faecalis CG110/gfp/pRE25*, a Tool for Monitoring Horizontal Gene Transfer in Complex Microbial Ecosystems,” FEMS Microbiology Letters, Vol. 313, No. 2, pp. 111-119. doi:10.1111/j.1574-6968.2010.02

