Open Journal of Genetics
Vol.4 No.1(2014), Article ID:43466,10 pages DOI:10.4236/ojgen.2014.41005
Hybrid Vigour and Genetic Control of Some Quantitative Traits of Tomato (Solanum lycopersicum L.)
Chinedozi Amaefula1, Christian U. Agbo1*, Godson Emeka Nwofia2
1Department of Crop Science, University of Nigeria, Nsukka, Nigeria
2Department of Agronomy, Micheal Okpara University of Agriculture, Umudike, Nigeria
Email: *christian.agbo@unn.edu.ng
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/


Received 8 January 2014; revised 7 February 2014; accepted 3 March 2014
ABSTRACT
Parental lines, F1 generation of domesticated tomato (Solanum lycopersicum L.) and Lycopersicon pimpinellifolium L. were evaluated at Department of Crop Science, University of Nigeria, Nsukka for improvement in their quantitative traits. The experiment was laid out in a randomized complete block design with three replications. Data were collected on the quantitative traits and fruit yield. Better Parent Heterosis (BPH), genetic variances, gene effects and heritability of the traits were estimated for the hybrids. BPH result showed that the Wild × Petomech cross had the highest positive BPH of 358.36% in fruit yield. The highest negative BPH of −95.59% was recorded for the hybrid, Wild × Grosso in average fruit weight while the hybrid, Insulata × Grosso had the lowest negative BPH of −16.27% in average fruit weight. Additive gene action and additive × additive gene action (aa) were significantly in control of three crosses, W × P, W × In and W × G in fruit yield. Additive variance was higher than dominance variance in fruit yield for all the hybrids with wild as one of its parents. Again, hybrids with wild as one of its parent as, W × G, W × In, and W × P had the highest narrow sense heritability in fruit yield (59.15%, 51.69%, 59.88%, respectively). High level of epistasis controlled some of the quantitative traits and hybridization evidenced by the result was effective in developing new tomato cultivars with positive heterotic effects in fruit yield.
Keywords:Gene Effect; Genetic Variance; Heritability; Heterosis; Hybrids

1. Introduction
The domestication and improvement of crops through breeding have been highly effective in concentrating allelic variation that confers useful characteristics for cultivation and consumption [1] . The objective of hybridization in breeding self-pollinated crops, is to combine in a single genotype genes that are found in two or more different genotypes [2] . The ability to use a particular wild relative depends on the recovery of progeny from the initial and subsequent crosses of tomato with the wild source, although all species can be crossed with tomato, the ease of success varies greatly [1] .
Hybrid tomato usually produces higher yield, they generally mature earlier and more uniformly [3] . Hybrid plants are usually heavy producers, and they combine the character of the parent plants. Many hybrids have better fruit quality and disease resistance. Resistance genotype should also possess other desirable economic traits to make them viable at commercial level [4] . Previous studies have suggested that increasing genetic distances (variability) between parents, increases heterosis [5] [6] .
Choudhary et al. [7] emphasized the effective utilization of heterosis to step up tomato production. Heterosis can be expressed when the parents of a hybrid have different alleles at a locus and there is some level of dominance or epistasis among the alleles [8] . It has been suggested that plant yield is a multiplicative trait that integrates variation from several other traits and therefore it may be expected that the trait would exhibit higher level of heterosis [9] . Allard [2] observed that the beneficial effect of crosses appear immediately in the F1 exhibiting heterosis. When parents differ considerably in type, the yields of the hybrids will be, with fewer exceptions, substantially greater than those of the better parent [2] [10] . The increased yield of hybrids could be as a result of high yielding parents selected for hybridization [11] . Sharma et al. [12] observed negative better parent heterosis in average fruit weight.
According to Mather & Jinks [13] , generation mean analysis is a useful technique that gives the estimation of main genetic effects such as additive, dominance and their allelic interactions involved in the expression of quantitative traits. The prevalence of any of the genetic effects will largely determine an effective breeding method for further development of new cultivars. El-Agamy et al. [14] had suggested maximum progress in new cultivar development using pedigree selection in traits where non-additive gene effect is prevalent where as hybridization will be effective in traits dominated by dominance and epistatic gene effects.
The need to develop tomato genotypes that will replace the existing exotic and landrace types that are either not adaptable or poor in quality motivated this study. The objective of this research was to develop new tomato genotypes expressing heterosis in fruit yield and quality and investigate the genetic control of the main quantitative traits controlling fruit yield in tomato. This information will be useful in the development of new cultivars with improved fruit quality and tolerant to high temperature, rainfall and disease infestations.
2. Materials and Method
The experimental materials used for the study were three parental lines of domesticated tomato (Solanum lycopersicum L.) namely; Petomech, Grosso and Insulata obtained from Naples, Italy and Lycopersicon pimpinellifolium (the wild parent) obtained from Mbu in Isi-Uzo Local Government Area of Enugu state, Nigeria. A 4 × 4 diallel analysis using Griffing’s [15] model 1 method 2 was employed to produce 6 F1 hybrids. The parental lines and the F1 hybrids were evaluated at the teaching and research farm of the Department of Crop Science, University of Nigeria, Nsukka. The experiment was laid out in a randomized complete block design with three replications. Each replication and plot was separated by a 1 meter path way. Well cured poultry manure was broadcast at the rate of 10 tonnes/ha a week before transplanting. Transplanting was done at one month after planting with a spacing of 1 m × 0.6 m. NPK 20:10:10 fertilizer was applied at the rate of 300 kg/ha one month after transplanting. Weeding and all cultural practices were carried out as at when due. The parents were evaluated along with the hybrids on the following traits; number of flowers/truss (NFT), number of trusses/plant (NTP), number of fruits/truss (NFRT), number of fruit/plant (NFP), fruit yield (FY), average fruit weight (AFW). Heteriosis was estimated as better parent heterosis (BPH) as put forth by [2] [16] as follows:

where ![]() is the mean of hybrid,
is the mean of hybrid, ![]() is the mean of the better parent.
is the mean of the better parent.
Test of significance was done as described by Kumar et al. [17] :

t = t tabulated at 5% probability;
r = number of replications;
me = error mean square;
2 = a constant.
Components of the generation means were evaluated using Hayman [18] model as explained by Singh & Chaudhary [19] as follows:
![]()

![]()

![]()
t value of effect = 
a = additive mean;
d = dominance effect;
aa = additive × additive;
ad = additive by dominance;
dd = dominance × dominance;
![]() = mean of backcross to parent 1;
= mean of backcross to parent 1;
![]() = mean of backcross to parent 2;
= mean of backcross to parent 2;
![]() = mean of parent 1;
= mean of parent 1;
![]() = mean of parent 2;
= mean of parent 2;
![]() = mean of First filial generation;
= mean of First filial generation;
![]() = of mean second filial generation;
= of mean second filial generation;
SE = standard error.
The estimate of the genetic variances of the quantitative traits was determined using the variance estimate method as described [16] [20] .
![]()

![]()




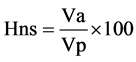
whereVe = Environmental Variance;
Va = Additive Variance;
Vd = Dominance Variance;
Vp = Phenotypic Variance;
Vg = Genotypic Variance;
Hb = Broad Sense Heritability;
Hns = Narrow Sense Heritability.
The variances of the parental lines, F1, F2, BC1 and BC2 population were used in determining the additive variance, dominance variance, genotypic variance, phenotypic variance, environmental variance, and heritability.
3. Results
Estimates of Better Parent Heterosis (BPH) of the agronomic, yield, yield component traits showed that, negative BPH was recorded in number of flowers/truss for all the crosses with W × G having the lowest negative value of –35.6% while G × P had lower negative BPH value of −4.82% (Table 1). The cross, In × G had higher BPH of 19.42% and 14.56% in number of trusses/plant, and fruits/truss, respectively than all the hybrids. The hybrid, In × P had the lowest negative BPH in number of fruits/plant (−23.52%). The cross, W × P had the highest positive BPH of 358.36% in fruit yield. All the hybrids had negative BPH in average fruit weight. The highest negative BPH of −95.59% was recorded for the hybrid, W × G for average fruit weight while the hybrid In × G had the lowest negative average fruit weight of −16.27%.
The result of the genetic effects of the agronomic, yield and yield traits of the tomato varieties studied showed that, significant additive gene effect was shown in W × P, W × In and W × G for number of flowers/truss (Table 2(a)). The cross, W × P, had significant ad gene action in number of flowers/truss. Dominance × dominance (dd) gene action was recorded in In × G, In × P, G × P and W × In in number of flowers /truss. Additive gene action was significant in the crosses, In × G, W × P, W × In and W × G (Table 2(a)). The aa gene effect was also significant in In × G, In × P, W × P, W × In and W × G in number of trusses/plant. The crosses, W × P, W × In, W × G had significant additive gene effects on the trait (Table 2(a)). There was a dd gene effect in all the crosses with the exception of In × P. Significant ad effect showed in W × P in number of fruits/truss. It was observed
Table 1 . Estimates of the Better Parent Heterosis (BPH) of the agronomic, yield and yield component traits of the F1 hybrids of tomatoes used for the study.
NFT = number of flowers/truss; NTP = number of trusses/plant; NFRT = number of fruits/truss; NFP = number of fruits/plant; FY = fruit yield; AFW = average fruit weight; In × P = Insulata × Petomech; In × G = Insulata × Grosso; W × G = wild × Grosso; W × In = Wild × Insulata; W × P = Wild × Petomech; G × P = Grosso × Petomech; cd = critical difference.
Table 2. (a & b) Gene effects on the agronomic, yield and yield traits of the crosses used in the diallel analysis of the tomato crosses used for the study.
NFT = number of flowers/truss; NTP = number of trusses/plant; NFRT = number of fruits/truss; m = F2 mean; a = additive effect; d = dominant effect; aa = additive × additive effect; ad = additive x dominant effect; dd = dominance × dominance effect; In × P = Insulata × Petomech; In × G= Insulata × Grosso; W × G = wild × Grosso; W × In = Wild × Insulata; W × P = Wild × Petomech; G × P = Grosso × Petomech.
Where: * = sig. at 5% level; NFP = number of fruits/plant; FY = fruit yield; AFW = average fruit weight; m = F2 mean; a = additive effect; d = dominant effect; aa = additive × additive effect; ad = additive × dominant effect; dd = dominance × dominance effect; In × P = Insulata × Petomech; In × G = Insulata × Grosso; W × G = wild × Grosso; W × In = Wild × Insulata; W × P = Wild × Petomech; G × P = Grosso × Petomech.
that additive gene action was significant in four cross combinations including: In × G, W × P, W × In and W × G in number of fruits/plant (Table 2(b)). Significant aa gene effect was recorded in In × P, W × P and W × In. Also, the crosses, In × P, W × P, W × In and W × G had significant ad effects in number of fruits/plant. In average fruit weight, the cross, G × P had significant additive gene effect while aa gene effect was significant in W × G cross for average fruit weight (Table 2(b)). Additive gene action was found to be significant in three crosses, W × P, W × In and W × G (Table 2(b)). Additive × additive aa gene action was also significant in W × P, W × In and W × G crosses.
Estimates of Variance Components, Broad and Narrow Sense Heritability
A decomposition of phenotypic variance into additive, dominant and component were carried out for the different crosses (Tables 3-8). Additive variance was higher than dominance variance in fruit yield for all the hybrids having the wild as one of its parents that is W × In (7925.091), W × In (3610.39) and W × P (9728.06). Dominance variance was higher than the additive variance in fruit yield for hybrids of two exotic parents such as In × P (2725.24), In × G (3676.97) and G × P (2272.85). Fruit yield had the highest environmental variance than all the traits in all the hybrids studied. Hybrids with wild as one of its parents had higher narrow sense heritability in fruit yield (W × G, 59.15%; W × In, 51.69%, and W × P; 59.88%). The other hybrids had low narrow sense (<50) heritability in fruit yield.
4. Discussion
The magnitude of heterosis depends on the accumulation of favourable dominant alleles in the F1 population.
Table 3. Estimates of the variance components, broad and narrow sense heritability of the cross between Insulata and Petomech (In × P).
Ve = environmental variance; Va = additive variance; Vd = dominance variance; VP = phenotypic variance; Vg = genotypic variance; Hbs = broad sense heritability; Hns = narrow sense heritability; NFT = number of flowers/truss; NTP = number of trusses/plant; NFRT = number of fruits/truss; NFP = number of fruits/plant; FY = fruit yield; AFW = average fruit weight.
Table 4. Estimates of the variance components, broad and narrow sense heritability of the cross between Insulata and Grosso (In × G).
Ve = environmental variance; Va = additive variance; Vd = dominance variance; VP = phenotypic variance; Vg = genotypic variance; Hbs = broad sense heritability; Hns = narrow sense heritability; NFT = number of flowers/truss; NTP = number of trusses/plant; NFRT = number of fruits/truss; NFP = number of fruits/plant; FY = fruit yield; AFW = average fruit weight.
Table 5. Estimates of the variance components, broad and narrow sense heritability of the cross between Wild and Grosso (W × G).
Ve = environmental variance; Va = additive variance; Vd = dominance variance; VP = phenotypic variance; Vg = genotypic variance; Hbs = broad sense heritability; Hns = narrow sense heritability; NFT = number of flowers/truss; NTP = number of trusses/plant; NFRT = number of fruits/truss; NFP = number of fruits/plant; FY = fruit yield; AFW = average fruit weight.
Table 6. Estimates of the variance components, broad and narrow sense heritability of the cross between Wild and Insulata (W × In).
Ve = environmental variance; Va = additive variance; Vd = dominance variance; VP = phenotypic variance; Vg = genotypic variance; Hbs = broad sense heritability; Hns = narrow sense heritability; NFT = number of flowers/truss; NTP = number of trusses/plant; NFRT = number of fruits/truss; NFP = number of fruits/plant; FY = fruit yield; AFW = average fruit weight.
Table 7. Estimates of the variance components, broad and narrow sense heritability of the cross between Wild and Petomech (W × P).
Ve = environmental variance; Va = additive variance; Vd = dominance variance; VP = phenotypic variance; Vg = genotypic variance; Hbs = broad sense heritability; Hns = narrow sense heritability; NFT = number of flowers/truss; NTP = number of trusses/plant; NFRT = number of fruits/truss; NFP = number of fruits/plant; FY = fruit yield; AFW = average fruit weight.
Negative BPH that occurred in all the crosses in number of flowers/truss, and number of fruits/plant could be as a result of long distance in the traits between the exotic and the wild parent. However, the results are in agreement with the findings of Sharma et al. [12] who observed negative heterosis in number of fruits/plant in tomato. In fruit yield, high positive BPH was recorded in all the crosses having the wild as the mother plant (Pistillate parent) probably because the wild had transferred traits for high yield to such crosses. When parents differ considerably in type, the yields of the hybrids will be, with fewer exceptions, substantially greater than those of the
Table 8 . Estimates of the variance components, broad and narrow sense heritability of the cross Grosso × Petomech (G × P).
Ve = environmental variance; Va = additive variance; Vd = dominance variance; VP = phenotypic variance; Vg = genotypic variance; Hbs = broad sense heritability; Hns = narrow sense heritability; NFT = number of flowers/truss; NTP = number of trusses/plant; NFRT = number of fruits/truss; NFP = number of fruits/plant; FY = fruit yield; AFW = average fruit weight.
better parent [2] [10] . Tolerance of the wild traits to high temperature and rainfall pattern of the study area coupled with high components of yield are good indicators of higher yield in the wild variety. Also the increased yield of hybrids could be as a result of high yielding parents selected for hybridization [11] . Hence, the dominance of such traits of the wild in all the crosses where the wild was the mother parent indicated the presence of maternal effect and BPH for fruit yield. This result is in conformity with the report of Dharmatti et al. [21] who showed a positive BPH for fruit yield in tomato. Earlier, Sharma et al. [12] had reported a negative BPH in fruit yield of tomato hybrids. The negative BPH recorded in the average fruit weight for all the crosses studied showed that none of the crosses had fruit weight that was bigger than the better parent. This could be attributed to the dominating effect of the small fruit size over the larger fruit size. This is in agreement with Sharma et al. [12] who observed negative BPH in average tomato fruit weight.
Narrow sense heritability is of great importance to the breeder. This is because it is the ratio of additive variance to total variance. Additive variance is the variance that causes resemblance among relatives [20] . The high narrow sense heritability (>50) recorded in fruit yield of W × G, W × In and W × P showed that these traits are highly heritable and should be selected for further studies in those crosses. This result is in conformity to the findings of Ghosh et al. [22] who recorded high heritability and high genetic advances in trusses/plant, fruits/plant, branches/plant, fruits/truss, fruit weight, and yield/plant of tomato hybrids. Wide levels of variation in broad sense heritability and narrow sense heritability in number of trusses/plant, fruit yield and average fruit weight in the crosses involving the exotic alone, In × P and G × P as well as exotic by wild is suggestive of higher environmental influence in the performance of such traits than other ones.
Phenotypic variance was higher than the genotypic variance in all the traits showing that there was an interaction of the traits with the environment. However, the low environmental variance in most of the traits suggests that the differences observed were mainly genetic. Traits high in narrow sense heritability and genetic variance indicated that they are controlled mainly by additive genes that are heritable and thus transferred from one generation to another. Such additive inheritance has been reported by Causse et al. [23] in some traits in hybrids between large-fruited and cherry tomato fruit lines.
Positive and significant additive gene effects occurred in all the crosses that had the wild as one of its parent in number of flowers/truss, trusses/plant, fruits/truss, and fruits/plant. These traits are therefore highly heritable. This agrees with Gamble [24] that gene effect is positive if better performing inbreds are used as P1. In number of fruits/plant, aa and ad were significant in crosses between the wild and an exotic. Dominance × dominance effects were significant in all the crosses except in number of fruits/truss in In × P. This result is in agreement with Zdravkovic et al. [25] who reported dd interaction in fruit weight. In fruit yield, additive, aa were significant in all the crosses between wild and an exotic parent. This showed that these traits can be fixed for possible selection of promising genotypes at early generation. Average fruit weight that showed significant epistatic, additive × additive effect in only a cross (W × G) with wild as parent is suggestive of the expression of linkage drag from the wild variety small fruit size that dominated the F1. The wild variety is intended to transfer genes for resistance to disease, adaptability to environmental conditions and high fruit number/plant. It goes on to transfer as well as the genes that reduces the fruit size and that quality affects the fruit size of the F1 not minding the fruit size of the better parent, even though the F1 in most crosses gave higher fruit yield. The prevalence epistatic, additive × additive, and additive × dominance gene control in the crosses with wild as a parent in number of fruits/plant could be the expression of high level of fruit number on a plant in the wild which tends to dominate the exotic variety.
5. Conclusion
Better parent heterosis which is of great importance to farmers was found to be higher in crosses having the wild as the pistilate parent, (W × G, W × P and W × In) for fruit yield. Hence, our findings show that the wild tomato variety is a good donor of genes for improvement of quantitative traits and yield in tomato. Also high narrow sense heritability was recorded in these hybrids for these traits. High narrow sense heritability and genetic variance observed in some traits indicated that they are controlled mainly by additive genes that are heritable and thus transferred from one generation to another. The high level of epistasis in the control of number of fruits/plant in those crosses with hybrid vigour in fruit yield indicated that the trait was very important in determining high yield and hybridization was effective in developing new tomato cultivars with heterotic effects in fruit yield.
References
- Osborn, T.C., Chad, K., Elaine, G. and Carl, J.B. (2007) Insight and Innovations from Wide Crosses: Examples from Canola and Tomato. Crop Science Society of America, Madison.
- Allard, R.W. (1960) Principles of Plant Breeding. John Willey and Sons Inc., New York.
- Shankara, N., Joep, V.D.J., Marja, D.G., Martin, H. and Barbara, V.D. (2005) Cultivation of Tomato: Production Processing and Marketing. Agromisa Foundation, Wageningen, 63-64.
- Kumar, Y.K.H., Patil, S.S., Dharmatti, P.R., Byadagi, A.S., Kajjidoni, S.T. and Patil, R.H. (2009) Estimation of Heterosis for Topovirius Resistance in Tomato. Karnataka Journal of Agricultural Sciences, 22, 1073-1075.
- Moll, R.H., Lonnquist, J.H., Fortuno, J.V. and Johnson, E.C. (1965) The Relationship of Heterosis and Genetic Divergence in Maize. Genet, 52, 139-144.
- Melchinger, A.E. (1999) Genetic Diversity and Heterosis. In: Coors, J.G. and Stuab, J.E., Eds., The Genetics and Exploitation of Heterosis and Crop Plants, Crop Science Society of America, Madison, 99-118.
- Choudhary, B., Punia, R.S. and Sangha, H.S. (1965) Manifestation of Hybrid Vigour in F1 and Its Correlation in F2 Generations of Tomato (Lycopersicon esculentum Mill). Indian Journal of Horticulture, 22, 55-59.
- Falconer, D.S. and Mackay, T.F.C. (1996) Introduction to Quantitative Genetics. 4th Edition, Prentice Hall, Essex.
- William, W. (1959) The Heterosis and the Genetics of Complex Characters. Nature, 184, 527-530.http://dx.doi.org/10.1038/184527a0
- Hossain, M.A., Khan, M.R.I. and Shaikh, M.A.Q. (1982) Morphological and Genetics in the Hybrid Derivatives of Oxheart and Anobik Tomato Cross. Bangladesh Journal of Botany, 11, 37-54.
- Courtney, W.H. and Peirce, L.C. (1979) Parent Selection Information Based on Morphological Trait. HortScience, 14, 458.
- Sharma, D.K., Chaudhary, D.R. and Pandey, D.P. (2001) Studies on Hybrid Vigour in Tomato (Lycopersicon esculentum Mill.). Haryana Journal of Horticultural Sciences, 30, 236-238.
- Mather, K. and Jinks, J.I. (1977) Introduction to Biometrical Genetics. Chapman and Hall, London.http://dx.doi.org/10.1007/978-94-009-5787-9
- El-Agamy, A.L., Eweida, M.H.T. and Okaz, A.M. (1975) Studies of Gene Action for Agronomic Characters in Cotton. Al-Azhan Agricultural Research Bulletin, 9, 3-10.
- Griffings, B. (1956) Concept of General and Specific Combining Ability in Selation to Diallel Crossing Systems. Australian Journal of Biological Sciences, 9, 463-493.
- Uguru, M.I. (2005) Crop Genetics and Breeding. 2nd Edition, Ephrata Press, Nsukka.
- Kumar, A., Mishra, V.K., Vyas, R.P. and Singh, V. (2011) Heterosis and Combining Ability Analysis in Bread Wheat (Triticum aestivum). Journal of Plant Breeding and Crop Science, 3, 209-217.
- Hayman, B.I. (1958) The Separation of Epistatic from Additive and Dominance Variation in Generation Means. Heredity, 12, 371-390. http://dx.doi.org/10.1038/hdy.1958.36
- Singh, R.K. and Chaudhary, B.D. (1985) Biometrical Methods in Quantitative Genetic Analysis. Kalyani Publishers, New Delhi.
- Acquaah, G. (2007) Principles of Plant Genetics and Breeding. Blackwell Publishing, Oxford.
- Dharmatti, P.R., Kulkarni, G.P. and Patil, R.V. (2006) Heterosis for Yield and Bacterial Wilt Incidence in Tomato. Journal of Asian Horticulture, 2, 151-154.
- Ghosh, K.P., Islam, A.K.M.A., Main, M.A.K. and Hossain, M.M. (2010) Variability and Character Association in F2 Segregating Population at Different Commercial Hybrids of Tomato (Solanum lycopersicon L.). Journal of Applied Sciences and Environmental Management, 14, 91-95.
- Causse, M., Buret, M., Robini, K. and Verschave, P. (2003) Inheritance of Nutritional and Sensory Quality Traits in Fresh Market Tomato and Relation to Consumer Preferences. Journal of Food Science, 68, 2342-2350.http://dx.doi.org/10.1111/j.1365-2621.2003.tb05770.x
- Gamble, E.E. (1962) Gene Effects in Corn (Zea mays L.) Relative Importance of Gene Effects for Plant Height and Certain Components Attributes of Yield. Canadian Journal of Plant Science, 42, 349-358.http://dx.doi.org/10.4141/cjps62-049
- Zdravkovic, J., Pavlovic, N., Girek, Z., Milka, B.J., Savic, D., Zdravkovic, M. and Cvikic, D. (2011) Generation Mean Analysis of Yield Component and Yield in Tomato (Lycopersicon esculentum Mill.). Pakistan Journal of Botany, 43, 1575-1580.
NOTES

*Corresponding author.


