American Journal of Molecular Biology
Vol. 2 No. 4 (2012) , Article ID: 23951 , 10 pages DOI:10.4236/ajmb.2012.24035
Biocatalytic production of chitosan polymers from shrimp shells, using a recombinant enzyme produced by Pichia pastoris
![]()
1Universidade Federal do Rio de Janeiro, Instituto de Química, Rio de Janeiro, Brazil
2Universidade Federal do Rio de Janeiro, Instituto de Macromoléculas Professora Eloisa Mano, Rio de Janeiro, Brazil
Email: emda@iq.ufrj.br
Received 4 August 2012; revised 4 September 2012; accepted 19 September 2012
Keywords: Chitin Deacetylase; Chitosan; Chitin, Pichia pastoris; Deacetylation Degree
ABSTRACT
Chitosan has a unique chemical structure with high charge density, reactive hydroxyl and amino groups, and extensive hydrogen bonding. Chitin deacetylase (EC 3.5.1.41) catalyzes the hydrolysis of the N-acetamido groups of N-acetyl-D-glucosamine residues in chitin, converting it to chitosan and releasing acetate. The entire ORF of the CDA2 gene encoding one of the two isoforms of chitin deacetylase from Saccharomyces cerevisiae was cloned in Pichia pastoris. The Tg (Cda2-6xHis)p was expressed at high levels as a soluble intracellular protein after induction of the recombinant yeast culture with methanol, and purified using nickel-nitrilotriacetic acid chelate affinity chromatography, resulting in a protein preparation with a purity of >98% and an overall yield of 79%. Chitin deacetylase activity was measured by a colorimetric method based on the (-phthalaldehyde reagent, which detects primary amines remaining in chitinous substrate after acetate release. The recombinant enzyme could deacetylate chitin, chitobiose, chitotriose and chitotetraose, with an optimum temperature of 50˚C and pH 8.0, determined using oligochitosaccharides as the substrates. The recombinant protein was also able to deacetylate its solid natural substrate, shrimp chitin, to a limited extent, producing chitosan with a degree of acetylation (DA) of 89% as determined by Fourier transform infrared spectroscopy. The degree of deacetylation was increased by pre-hydrolysis of crystalline shrimp chitin by chitinases, which increased the deacetylation ratio triggered by chitin deacetylase, producing chito-oligosaccharides with a degree of acetylation of 33%. The results described here open the possibility to use the rCda2p, combined with chitinases, for biocatalytic conversion of chitin to chitosan with controlled degrees of deacetylation. We show herein that the crystalline chitin form can be cleanly produced in virtually quantitative yield if a combined and sequential enzyme treatment is performed.
1. INTRODUCTION
Chitin is an unbranched polysaccharide composed of β- linked 2-acetamido-2-deoxy-D-glucose (N-acetyl-D-glucosamine) units found in the shells of crustaceans, cuticles of insects, cell walls of fungi, and in a number of other lower plants and animals [1]. In nature, 10 gigatons of chitin is synthesized and degraded each year. The chitin polymer displays a 0.9% degree of acetylation, 7% nitrogen content, and nitrogen/carbon ratio of 0.146 [2].
Chitin has many useful biological properties such as biocompatibility, biodegradability, hemostatic activity, and wound-healing property, but its use has been limited by its low solubility in water and most common organic solvents [3]. However, chitosan, a partially deacetylated form of chitin composed by N-acetyl-D-glucosamine and N-D-glucosamine units that are randomly or block-distributed throughout the biopolymer chain, is readily soluble in dilute acids, rendering it more accessible for utilization and chemical reactions. Chitosan has been used in several biomedical applications, in orthopedic tissue engineering [4], wound dressing and artificial skin, personal-care products [5], and for drug delivery [6].
Deacetylation of chitin to chitosan is usually achieved by thermo-chemical treatment using alkali solutions at high temperatures for extended periods of time. In the heterogeneous alkaline method, chitin is treated with a hot (70˚C - 150˚C), concentrated solution of NaOH (10% - 60%), and 5 to 45 mL of alkaline solution per gram of chitin. Chitosan prepared by this method is produced as an insoluble precipitate in a ~85% - 93% deacetylated form. Thermo-chemical methods have three critical disadvantages: 1) They consume considerable amounts of energy; 2) They waste a large amount of concentrated alkaline solution, resulting in an increase in the level of environmental pollution; and 3) They lead to products with a broad range of molecular weights and a heterogeneous extent of deacetylation [7]. A green alternative for chitosan production is the enzymatic deacetylation of chitin using chitin deacetylase under mild conditions [5], giving a product with more homogeneous characteristics.
Chitin deacetylase (CDA-EC 3.5.1.41) catalyzes the deacetylation of N-acetyl-D-glucosamine residues in chitin, and is a member of the carbohydrate esterase family 4 (CE-4s), as defined in the CAZY database (www.cazy.org) [8]. This enzyme plays a role in cell-wall biosynthesis in Mucor rouxii and Absidia coerulea [5]; it is suggested to participate in plant-pathogen interactions to promote plant invasion in Colletotrichum lindemuthianum and Aspergillus nidulans [5]; and to be essential to ascospore cell-wall rigidity and resistance against lytic enzymes in Saccharomyces cerevisiae [5].
The genes encoding chitin deacetylase from M. rouxii, Rhizopus circinans, S. cerevisiae and C. lindemuthianum have been cloned and characterized [5,9] and expressed in Escherichia coli [10] and in Pichia pastoris [11].
Two chitin deacetylase genes, namely CDA1 and CDA2, have been identified in S. cerevisiae. Comparison of the deduced amino acid sequence of Cda1p (284 amino acids, 34.6 kDa) and Cda2p (274 amino acids, 35.6 kDa) showed an identity of 57% and a similarity of 72% throughout their entire amino acid sequence, with a gap of 10 amino acids starting at position 39 of Cda1p [12]. Cda2p is a glycoprotein with an apparent molecular mass of approximately 43 kDa and a carbohydrate content of approximately 18% by weight. The purified enzyme has an optimum pH of 8.0 and an optimum temperature of activity at 50˚C, requires at least two N-acetyl-D-glucosamine residues (chitobiose) for catalysis, and is partially inhibited by acetate [13].
The mode of action of CDA from M. rouxii has been investigated on a ~32% randomly deacetylated watersoluble chitosan substrate, with an average polymerizetion degree of 30. Using 1Hand 13C-NMR spectroscopy, it was found that the enzyme hydrolysed the acetyl groups of the substrate according to a multiple-attack mechanism with a degree of multiple attack of three. This is the maximum number of successive deacetylations that could be achieved by the enzyme because the maximum number of the consecutive N-acetyl-D-glucosamine residues that were found in this substrate polymer was three [14].
The enzymes are highly homologous and, furthermore, there is a universal conserved region that exhibits a significant similarity to the rhizobial nodulation proteins (NodB proteins), certain regions in microbial acetyl xylan esterases, xylanases, peptidoglycan deacetylase and several uncharacterized open reading frames (ORFs) in Bacillus sp. This conserved region has been assigned as the NodB homology domain [15]. Multiple sequence alignment of chitin deacetylase family demonstrates that the sequences contain five well conserved catalytic motifs which make up the active site of the deacetylase domain in the CE-4 family [16]. The five catalytic motifs cover several conserved histidine and aspartic acid residues, which were similarly observed in other CE-4 family members such as S. pneumoniae peptidoglycan deacetylase and S. lividans xylan esterase [17].
The production, characterization and application of CDA are a fundamental and vital area of current research, considering its potential and biotechnologically significant role in chitin deacetylation. Considering this, it is pertinent to search for novel extracellular CDA producers, which could result in markedly higher levels of the enzyme. The CDA thus obtained should preferably be stable under standard conditions of catalysis such as pH, temperature, etc., which may translate into higher yields and potentially lower cost of the end product.
The development of viable processes for the controlled hydrolysis of chitosan is attracting interest, because the functional and biological effects of these biopolymers are highly dependent upon their composition and molar mass [18].
Chitinases (E.C. 3.2.1.14) are capable of converting chitins and chitosans to low-molecular-mass products. They randomly hydrolyze β-1,4 bonds between the sugar units, and produce N-acetylglycosamine and N,N’- diacetylchitobiose as the sole reaction products. Chitinases from higher plants are classified in families 18 and 19, based on the similarity of their sequences to the glycosyl hydrolases [19]. These classes of enzymes can break up the crystalline chitin structure, and should increase the accessibility of deacetylation enzymes to the resultant chitosans.
In the present study, the ORF encoding the Saccharomyces cerevisiae CDA2 gene was cloned and expressed in Pichia pastoris. The recombinant protein was purified to homogeneity, and its catalytic activity was evaluated by following the formation of primary amine on natural or artificial chitin substrates using the o-phthalaldehyde reagent. Chitosans with 33% deacetylation can be produced by bioconversion, using a combination of enzymatic treatment of crystalline chitin by chitinases and chitin deacetylase.
2. MATERIALS AND METHODS
2.1. Strains, Plasmids and Supplies
Pichia pastoris strain X33, Escherichia coli strain DH5α, plasmids pPICZ A and pUC18, restriction enzymes, Platinum® Taq DNA Polymerase High Fidelity (Invitrogen), shrimp phosphatase, and customized primers were purchased from Invitrogen. T4 polynucleotide kinase was obtained from USB. T4 DNA ligase was purchased from Promega. Ni-NTA Superflow was obtained from Qiagen. The gel extraction kit was purchased from GE Healthcare Life Sciences, and the GelCode® 6xHis Protein Tag Stain Reagent kit from Pierce. Di-N-acetyl chitobiose, tri-N-acetyl chitotriose and tetra-N-acetyl chitotetraose were purchased from IsoSep. Shrimp crystalline chitin was prepared in our laboratory. All other chemicals used were of reagent grade. Media LB (10 g bacto tryptone, 50 g bacto yeast extract and 10 g NaCl in 1 L) and YPD (10 g yeast extract, 20 g peptone and 20 g glucose in 1 L) were used for routine growth and maintenance of the E. coli and P. pastoris strains, respectively. LB medium plus ampicillin (25 µg/mL) and YPD medium plus zeocin (100 µg/mL) were used for selection of E. coli and P. pastoris transformants, respectively. Buffered glycerol-complex medium (BMGY) containing, per liter, 10 g yeast extract, 20 g peptone, 100 mM potassium phosphate, 13.4 g yeast nitrogen base without amino acids, and 10 mL glycerol, pH 6.0 was used for growth of P. pastoris transformants. The buffered methanolcomplex medium (BMMY) contained all the same components as BMGY, except glycerol, which was replaced by 10% methanol per liter, and was used for induction of the heterologous protein expression.
2.2. Cloning of CDA2 into pPICZ A Expression Vector
Genomic DNA was extracted from S. cerevisiae strain W303-1A (Sambrook and Russell, 2001) and used as a DNA template for PCR amplification, using the set primers Cda2forward (5’-CTCGAGATGAGAATACA ACTAAATAC-3) and Cda2reverse (5’-GGGCCCGGA CAAGAATTCTTTTAT-3’) carrying restriction sites for XhoI and ApaI, respectively (boldface). PCR was performed using Taq DNA polymerase High Fidelity™ (Invitrogen) for 30 cycles at 92˚C for 10 s, 55˚C for 30 s and 68˚C for 4 min, and 1 cycle at 68˚C for 7 min. The 948 bp PCR product was phosphorylated using T4 polynucleotide kinase and cloned into SmaI-digested and dephosphorylated plasmid pUC18. The resultant plasmid (pEMDA-P2) was transformed into DH5α cells, and the transformants were selected in Luria Broth (BD™) plus ampicillin (25 µg/mL) plates. Plasmid DNA from transformants was digested with ApaI and XhoI restriction enzymes, and the ApaI/XhoI 948 bp fragment purified from agarose gel was cloned into ApaI/XhoI-digested and dephosphorylated plasmid pPICZ A. The ligation product was transformed into DH5α, and positive clones were selected in LB plus zeocin (25 µg/mL) plates. Plasmid pEMDA-Z2 (4246 bp), containing the alcohol oxidase (AOX1) promoter and the entire open reading frame of the CDA2 gene, in frame with c-myc epitope and poly-histidine (6xHis) tag coding sequences, was isolated (Figure 1). The plasmid was cut in the 5’ AOX1 region with BglII, and transformed into P. pastoris strain X33 by electroporation (1500 V, 25 μ FD and 200 Ω). Transformants were selected on YPD plus zeocin (100 µg/mL) plates, and the presence of the chromosomal integration of the CDA2 expression cassette was assessed by a confirmatory PCR using primers 5’-GACTGGTTCCAATTGACAAGC-3’ and 5’-GCAA ATGGCATTCTGACATCC-3’ (AOX1 primers), performed for 30 cycles at 95˚C for 1 min, 54˚C for 1 min and 72˚C for 1 min, and 1 cycle at 72˚C for 7 min.
2.3. Overexpression of the Chitin Deacetylase Fusion Protein
Pichia pastoris cells X33/EMDA-Z2 were inoculated in 25 ml of BMGY medium at 28˚C with vigorous shaking (250 rpm), cultured to saturation (OD600 nm of 6.0) and harvested by centrifugation. The pellet cell was resuspended in 100 mL of BMMY medium and the cells were allowed to grow for a further 48 h, with the addition of 0.5% (v/v) methanol after the first 24 h.
2.4. Purification of the Cda2p Recombinant Protein
After methanol induction, X33/EMDA-Z2 transformants
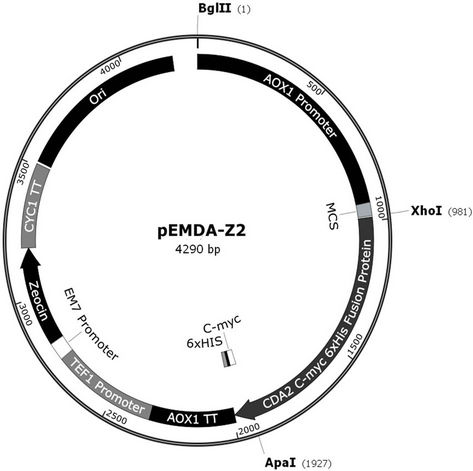
Figure 1. Recombinant map of pEMDA-Z2. PCR amplified S. cerevisiae CDA2 gene (0.945 kb) was inserted in pPICZA (3.3 kb) after Xho I/Apa I digestion.
were harvested by centrifugation (10 min at 4100 × g) and the pellet was resuspended in 10 mL of 0.05 M lysis buffer. The cell suspension was left in an ice bath for 30 min, homogenized for 1 min with an equal volume of glass beads in a Braun cell homogenizer (B. Braun Biotech), and then chilled for another 1 min in an ice bath. The crude extract was centrifuged at 12,000 × g (10 min at 4˚C) and the supernatant was loaded onto a 5 mL Ni-NTA Superflow column, previously equilibrated in 50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole pH 8.0 without lysozyme (1 mg/mL), and 1 mM PMSF, and allowed to interact for 18 h. Free proteins were washed with 100 mL of 50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole pH 8.0, and the bound proteins were eluted with 50 mL of 50 mM NaH2PO4, 300 mM NaCl, and 100 mM imidazole pH 8.0. The elution fractions were followed by measuring the optical density at 280 nm in a Beckman DU® 640 spectrophotometer (1 OD 280 nm = 1.866 mg/mL). The Cda2p pooled fraction was dialyzed against 10 mM Tris buffer pH 6.7 for 24 h, and filtered through a 0.22 μm-pore membrane.
Cda2p purification was evaluated by discontinuous denaturing SDS-polyacrylamide gel electrophoresis (12.5% acrylamide monomers in the running gel) performed according to Laemmli (1970) [20]. Gels were stained with Coomassie brilliant blue G-250. Polyhistidine-tagcontaining proteins were detected by using the Pierce® 6xHis Protein Tag Stain Reagent Set kit (Thermo Scientific) following the manufacturer’s recommendations.
2.5. Preparation of Chitinases
Chitinases were obtained from Vitis vinifera L. cv “Red Globe” and purified by fractionation in 80% (v/v) ammonium sulfate, followed by dialysis and filtration [21].
2.6. Chitin Deacetylase Activity Assays
Chitin deacetylase activity was estimated by using synthetic chito-oligosaccharides (di-N-acetyl chitobio-se, triN-acetyl chitotriose, and tetra-N-acetyl chitotetraose, IsoSep) or crystalline shrimp chitin, deproteinated, demineralized, and depigmented as described previously [22] as substrates [23]. In brief, 50 µg of each synthetic substrate or 1 mg of crystalline shrimp chitin was incubated with 0.3 and 2 U, respectively, of the purified recombinant chitin deacetylase in 25 mM phosphate buffer, pH 8.0, containing 1 mg/mL of BSA, for 1 h at 50˚C. The reaction was stopped by heating at 100˚C for 10 min. The amount of primary amine formed was determined spectrophotometrically after derivatization with o-phthalaldehyde (OPA). One unit of enzyme activity is defined as the amount of the enzyme required to produce 1 µmol of primary amine per minute under the above conditions.
2.7. Deacetylation of Shrimp Chitin Pre-Hydrolyzed by Chitinases
Ten milligrams of shrimp crystalline chitin was treated with 10 U of purified recombinant chitin deacetylase in 25 mM buffer, pH 8.0, containing 1 mg/mL of BSA (final volume 750 μL) for 1 h at 50˚C, as described above. Alternatively, crystalline chitin was pre-hydrolyzed by 6.0 U of purified chitinases in 50 mM sodium acetate buffer pH 3.0 at 42˚C for 50 min in a final volume of 20 mL. The reaction was stopped by heating at 100˚C for 10 min in sealed tubes, centrifuged (9000 g for 5 min), and the pellet was treated with Cda2p as described.
2.8. FTIR Spectroscopy Analysis of Chitosans
Fourier Transform Infrared (FTIR) spectra were recorded in a Perkin-Elmer spectrometer, model 1720 (Salem, MA, USA), with an accumulation of 20 scans, and a resolution of 2 cm–1. KBr, permanently maintained in an oven at 50˚C, was used to prepare transparent disks. Chitin samples and the resulting product obtained by enzyme treatment were thoroughly dried and carefully weighed (2 mg) before grinding with 100 mg·KBr. Spectral bands at 1420 cm–1 and 1320 cm–1 were taken as the reference and characteristic bands, respectively, and the absorbance ratio A1320/A1420 was calculated in order to determine DA [24].
3. RESULTS
3.1. Isolation and Sequencing of the CDA2 Gene
The YLR308W/CDA2 coding sequence was success-fully amplified by PCR using total genomic DNA from S. cerevisiae strain W303-1A as a template. The 948 bp PCR product was directly cycle-sequenced with ABI BigDye Terminator v. 3.1, using the primers Cda2forward and Cda2reverse in separate sequencing reactions on an ABI 3730 × l DNA Analyzer at the W. M. Keck Foundation Biotechnology Resource Laboratory (Yale University, USA). The sequences obtained were compared to the YLR308W/CDA2 sequence available at the Saccharomyces cerevisiae Genome Database (www.yeastgenome.org), by using the Clustal 3.0 program. The forward and reverse sequences covered 92.4% of the YLR308W/CDA2 ORF with only two mismatches, confirming that the amplified segment corresponded to the CDA2 gene (Figure 2).
3.2. Cloning of the CDA2 and Expression of the Heterologous Protein in P. Pastoris
The 948 bp amplified fragment was phosphorylated and inserted into SmaI-cut and dephosphorylated pUC18, creating pEMDA-P2 (3634 bp). The XhoI/ApaI EMDA-

Figure 2. Nucleotide sequence alignment of PCR product and CDA2 gene. Foward (CDA2-5) and reverse (CDA2-3) nucleotide sequences of the PCR product targeting yeast CDA2 gene were aligned with the CDA2 sequence available at Saccharomyces cerevisiae genome database (www.yeastgenome.org) using the Clustal 3.0 program.
P2-derived fragment was ligated to XhoI/ApaI-cut plasmid pICZA, producing the 4246 bp plasmid pEMDA-Z2 containing the entire YLR308W/CDA2 ORF under the control of AOX1 promoter and fused in frame with c-myc epitope/6xHis tag. BglII-cut pEMDA-Z2 was inserted into the host P. pastoris strain X33 by electroporation. Integration by homologous recombination of the expression cassette within the 5’ AOX1 chromosomal region was identified by the presence of the 1272 bp diagnostic fragment amplified by the AOX1 primers. Recombinant X33/EMDA-Z2 cells were grown in BMGY medium. The cells were transferred to BMMY medium [with 0.5% (v/v) methanol] and cultured for 48 h, with a new addition of methanol to a final concentration of 0.5% after 24 h to maintain induction. Under these conditions, SDS-PAGE analysis of the total cell protein revealed a protein band of 37 kDa, which appeared after 12 h, peaked at 48 h, and decreased in intensity after 60 h of induction; no corresponding band was present in the control cells (results not shown).
3.3. Purification of the Recombinant Cda2p
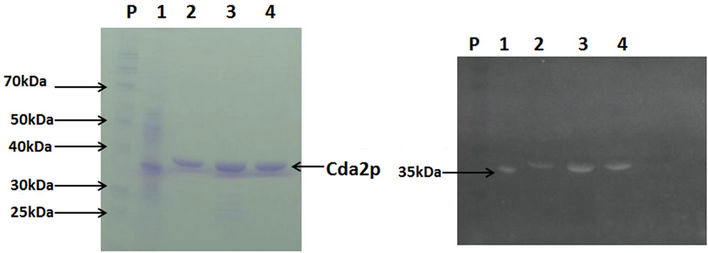
Recombinant CDA2p was purified to apparent homogeneity, as judged by the 12.5% SDS-PAGE, by a one-step procedure using nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography, with an overall yield of 79% and a purification factor of 11 (Table 1). The apparent molecular mass of the purified protein (37 kDa) (Figure 3(a)) corresponds to the expected mass of the recombinant peptide coded by the CDA2 ORF fused in frame with c-myc and 6XHis-tag coding sequences. The polyhistidine tail in the recombinant protein was confirmed by staining with the GelCode® 6xHis Protein Tag Stain reagent Set (Figure 3(b)).
The specific activity of the purified preparation (39.7 U/mg) was about 20-fold lower than the native Cda2p [23]. This difference can be attributed mainly to the use of (GlcNAc)6 to assay native Cda2p, which is a two-fold better substrate than (GlcNAc) 4 used in the present study [23] and perhaps to the absence of Co2+ in the reaction mix, since this metallic ion is known to enhance the chitin deacetylase activity [5].
3.4. Characterization of the Catalytic Activity of the Recombinant Cda2p
A novel colorimetric assay was developed for screening the Cda2p preparation activity, which is based on the specificity of the reaction of o-phthalaldehyde with primary amines formed upon deacetylation of the chitinous substrates, producing a colored product that was estimated at 340 nm.
Using N-acetylchitotetraose as a substrate and the o-phthalaldehyde reagent, the influence of pH (in the range from 7.0 to 10.0) and temperature (in the range from 30˚C to 80˚C) on the chitin deacetylase activity was evaluated. The optimum pH was 8.0, similar to those described in the literature [5]. The optimum temperature was 50˚C, at which the enzyme remained stable for at least 90 minutes (data not shown). Under these conditions, the primary amine was formed linearly up to 30 min, and all subsequent reactions were performed under these initial velocity conditions. The recombinant chitin deacetylase preparation exhibited the same pH and temperature reported in a previous study, considering the Cda2p expressed in both S. cerevisiae [25] and E. coli [10].
In order to test the deacetylation specificity of the recombinant chitin deacetylase, N-acetyl chito-oligosaccharides (chitobiose, chitotriose and chitotetraose) and crystalline shrimp chitin were used as substrates, and the amounts of primary amines formed were compared to those obtained with N-acetyl chitotetraose. The rCda2p deacetylated all the substrates tested, but the increase in
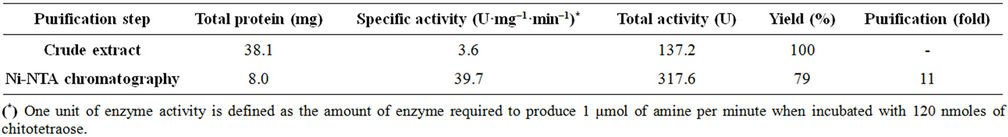
Table 1. Purification of recombinant chitin deacetylase.
 (a) (b)
(a) (b)
Figure 3. Electrophoretic analysis of the recombinant chitin deacetylase. Purified recombinant chitin deacetylase preparation was electrophoresed on a 12.5% SDS-PAGE. Protein bands were visualized by staining with Coomassie blue R (a) or by GelCode® 6xHis Protein Tag Stain reagent Set (b). lane P, protein molecular mass markers; lane 1, positive control lysate (35 kDa) from the 6xHis Protein Control Set (Pierce), lane 2, purified Cda2p following the purification.
chain length enhanced the effectiveness of the enzyme. The correlation between the structural features of the chitinous substrates and the extent of enzymatic deacetylation was clearly demonstrated when crystalline shrimp chitin was used as a substrate: an increased rate of deacetylation was observed, in comparison to the Nacetylchito-oligomers with 2, 3 and 4 units of polymerization (Table 2), as previously reported for the purified enzymes from S. cerevisiae [26] and from Scopulariopsis brevicaulis [5].
3.5. Biocatalytic Production of Chitosans
The deacetylation of shrimp chitin by the recombinant chitin deacetylase in 30 min led to a decrease of 5.1% in the degree of acetylation (Table 2). This result encouraged us to pursue the study of the deacetylation of Cda2p or its natural substrate. In a scaled-up experiment, 10 mg of crystalline shrimp chitin was treated by recombinant chitin deacetylase for 3 h, and the degree of acetylation (DA) of the chitin substrate was estimated by FTIR (Figure 4). In FTIR spectra, the amide I band was split into two components, attributed to the two types of amide H-bonds formed in high-molecular-weight a-chitin [27]. The amide II band at 1550 cm–1, attributed to the C-N bond stretching and to C-N-H bonds bending in secondary amides, was also present. From the spectral data, DA was estimated by using the calibration curve reported in the literature [28] and the absorbance ratios A1320/A1420 were determined. These ratios led to a DA of 100% and 89% for control and enzyme-deacetylated chitin samples, respectively. The chitinous product showed a DA of 89% after deacetylase treatment; although this succeeded well, it was necessary to increase the deacetylation. This was tested by pre-hydrolysis of crystalline shrimp chitin by chitinases, which increased the deacetylation ratio, triggered by chitin deacetylase, producing chito-oligosaccharides with a DA of 33% (Figure 5). The remaining major impediment to an enzymatic hydrolysis process is the extremely low hydrolytic susceptibility of the natural chitin substrate, due to its high degree of crystallinity. We showed herein for the first time that the crystalline chitin form can be cleanly produced in virtually quantitative yield if a combined and sequential enzyme treatment is performed.
4. DISCUSSION
A chitin deacetylase protein from S. cerevisiae was cloned and expressed in P. pastoris, and the recombinant enzyme was purified to homogeneity. The methyl-trophic yeast P. pastoris proved to be a suitable system for heterologous protein expression, because of, among other favorable properties, the existence of a strong and tightly regulated promoter from the alcohol oxidase 1 gene, AOX, which in combination with an optimal codon usage bias when expressing the CDA2 gene from S. cerevisiae, accommodated high expression levels of the Cda2
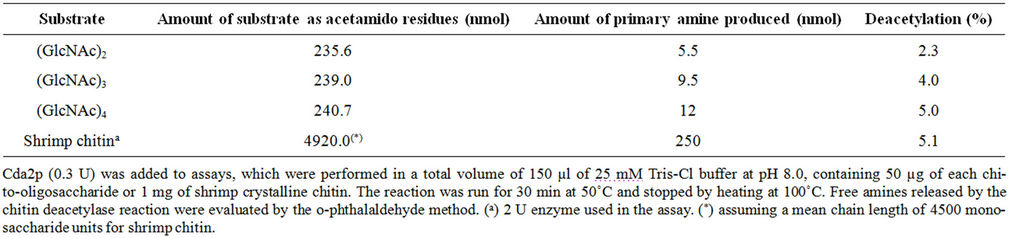
Table 2. Deacetylation of N-acetylchito-oligosaccharides and chitin by the recombinant Cda2 protein.

Figure 4. FT-IR spectra of chitin deacetylase products. (a) Shrimp crystalline chitin; (b) 10 mg of shrimp crystalline chitin treated by recombinant chitin deacetylase (0.3 U) for 3 h in 25 mM phosphate buffer, pH 8.0, containing 1 mg/mL of BSA at 50˚C.
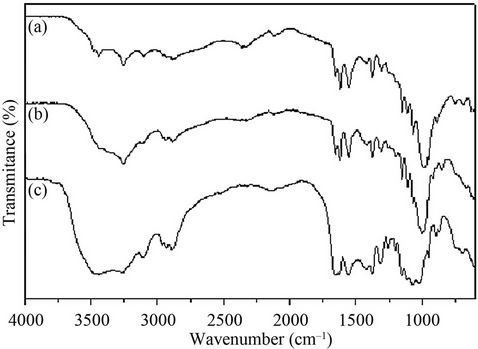
Figure 5. FT-IR spectra of chitosans produced by chitinase and chitin deacetylase treatment. (a) Shrimp crystalline chitin; (b) 10 mg of shrimp crystalline chitin treated by recombinant chitin deacetylase (2 U) for 1 h in 25 mM phosphate buffer, pH 8.0, containing 1 mg/mL of BSA at 50˚C; (c) 10 mg of shrimp crystalline chitin treated by purified chitinase (1.2 U) on 50 mM sodium acetate buffer pH 3.0 at 42˚C for 50 min, followed by treatment with purified recombinant chitin deacetylase (2 U) for 1 h.
protein. In addition, P. pastoris is transformed by integration of the expression cassette into the chromosome at a specific locus, to generate genetically stable Cda2p transformants [29]. Methanol-induced expression of CDA2 integrated into the P. pastoris genome allowed largescale production of a soluble 37 kDa cytosolic protein. This was compatible with the expected molecular mass for cda2 chitin deacetylase fused with c-myc epitope and polyhistidine (6xHis) tag recombinant protein, it was predicted to have 310 amino acids overall and a molecular mass of 35558.5 Da, assuming that the putative signal peptide (25 amino acids long) was processed.
The agreement with previous reports, the apparent molecular mass of the purified protein is close to 35 kDa, similar to the Cda2p expressed in Escherichia coli and to the N-deglycosylated enzyme form expressed in P. pastoris [13]. The results described here are also consistent with the expression of the enzyme in its N-deglycosylated form, as previously reported for the recombinant C. lindemuthianum chitin deacetylase expressed in P. pastoris [25]. Treatment of the purified protein in the polyacrylamide gel with thymol and sulfuric acid showed that it was not a glycosylated protein (results not shown) [30]. These results, however, differ from those observed for native enzymes that were purified in their glycosylated forms from different organisms such as C. lindemuthianum [31], M. rouxii [32], and S. cerevisiae [33].
Purification of the recombinant enzyme to apparent homogeneity (Figure 3(a)) was effectively achieved by a single-step chromatography procedure employing a Ni-NTA column, which exploits the 6xHis tag added to the recombinant Cda2p. The specific activity of the purified preparation (39.7 U/mg) was about 20-fold lower than that of native Cda2p [13]. This difference can be attributed mainly to the use of (GlcNAc)6 as a substrate to assay native Cda2p, which is a two-fold better substrate than the (GlcNAc)4 used in the present study [25]; and, perhaps, to the absence of Co2+ in the reaction mix, since this metallic ion is known to enhance the chitin deacetylase activity [5]. Notably, total loss of enzyme activity was observed after deglycosylation of Cda2p from S. cerevisiae [13], which could be restored by the addition of 1 mM CoCl2. Here, recombinant Cda2p was shown to be active in its deglycosylated form and in the absence of Co2+. The effect of Co2+ on the enzyme activity was not tested.
The recombinant chitin deacetylase preparation exhibited the same optimum pH (8.0) and temperature (50˚C) reported for the recombinant Cda2p expressed in S. cerevisiae [13] and in E. coli [10]. It seems that a high optimum temperature is a characteristic of this enzyme, since optimum temperatures in the range of 50˚C to 60˚C have been reported for chitin deacetylase from A. coerulea [34] and several other fungal enzymes [5]. Similarly, the optimum pH of most extracellular chitin deacetylases ranges from 7 to 12 [5]. Using N-acetylchitotetraose as a substrate, pH 8.0 and at 50˚C, the reaction ran under initial velocity conditions for up to 30 min, as judged by the quantification of the primary amine formed using οphthalaldehyde [35].
Although this reagent has been reported to be useful to determine the degree of deacetylation of chitosan of various molecular weights [36], to the best of our knowledge, this is the first use of this reagent to assay chitin deacetylase activity. Up to the present, assays for chitin deacetylase activity have been based on the release of radioactive acetate from glycol chitin radiolabeled in the N-acetyl group [13]; on quantification of acetate release using acetyl-CoA synthetase, citrate synthase, and malate dehydrogenase in a coupled system [25,33]; or on chemical detection of glucosamine residues with 3- methyl-2-benzothiazolone hydrazone after deamination with nitrous acid [26,37]. Therefore, the use of a method that employs readily available reagents could help to increase the interest of researchers in the study of chitin deacetylase.
The purified chitin deacetylase was active on N-acetylchito-oligosaccharides [(GlcNAc)2, (GlcNAc)3 and (GlcNAc)4] and on native-shrimp crystalline chitin (Table 2). The deacetylase activity increased with the degree of polymerization of the N-acetylchito-oligosaccharides, as previously reported for the enzyme purified from S. cerevisiae [13] and from Scopulariopsis brevicaulis [26]. Acting on tetra-N-acetyl chitotetraose, the enzyme showed maximum activity, reaching 5% DA (degree of deacetylation). The degree of deacetylation of (GlcNAc)4 by chitin deacetylase found in this study was lower than previously reported [13]. However, in this study, we used a shorter assay time (30 minutes) than the time used in the previous report (48 hours).
When crystalline shrimp chitin, the natural substrate, was hydrolyzed by chitin deacetylase, the degree of deacetylation obtained was 11%, which is low but still higher than several others reported in previous studies [26], and paves the way for the development of an efficient enzymatic process for production of chitosan with a controlled degree of deacetylation. This is an encourageing result, since the deacetylation level is higher than those reported by other studies [26,31,34]. It is well known that the high crystallinity of chitin microfibril, promoted by the hydrogen bond-stabilized packaging of chitin polymer, greatly impedes the access of the enzyme to the deacetylation reaction site in the chitin molecule. Pretreatment to destroy the chitin crystalline structure prior to addition of the enzyme seems to be desirable, in order to improve the deacetylation rate and produce novel chitosan polymers and oligomers.
Most of the other chitinous substrates without deacetylation cannot be affected by CDA, even though their particle size is very small [38]. It is thought that enzymatic deacetylation is profoundly affected by the physical properties of the substrate, such as crystallinity, degree of deacetylation, particle size, and origin.
The pre-hydrolysis of chitin by grape chitinases increased the degree of deacetylation to 33%. It seems that the preparation of chitinases undoes the interaction between the molecules of chitin obtained from shrimp. This type of α-chitin forms a dense packaging resulting from the antiparallel arrangement of the polymer chains, which favors the existence of numerous interand intrachain hydrogen links in the same and neighboring lamellae, forming a rigid and resistant structure, which could hinder access of the recombinant deacetylase to the acetyl groups of the chito-oligosaccharide [39].
Nowadays, the deacetylation of chitin to chitosan is usually achieved using a strong alkali at high temperatures for extended periods of time. This is an environmentally unsuitable process, as is the treatment with hydrolytic, non-specific enzymes, a difficult-to-control process that frequently gives a broad and heterogeneous range of products. Moreover, because the enzyme preparations that are used tend to be rather crude and are derived from different organisms, including fungi and plants, that are known to produce chitinolytic enzymes, there remains some doubt concerning which enzymes actually catalyze the hydrolysis reaction [40].
Biocatalytic conversion of chitosans using the purified forms of both chitinases and chitin deacetylase has been described in few studies [21]. Recently, several comercially available hydrolytic enzymes including lysozyme, cellulase, papain, pectinases, and hemicellulase were found to catalyze the cleavage of the glycoside bond in chitosan [26,41]. However, the use of enzymes with narrow specificity for the substrate (chitin), chitinases and chitin deacetylase, certainly opens a possible route to optimize the hydrolysis reactions controlling the production of chito-oligosaccharides.
The practicality of using this recombinant protein for biocatalytic conversion of chitin to chitosan, mainly when combined with chitinases, which in optimum conditions can produce chitosan oligomers with defined sizes and degrees of deacetylation for use in specific applications, was demonstrated. Preparation of chitin deacetylase from S. cerevisiae and grape chitinases could be a viable alternative for the production of industrialized chito-oligosaccharides, because enzymatic hydrolysis is rapid, the production conditions are mild, complicated production equipment is not required, and the cost of production is low.
5. ACKNOWLEDGEMENTS
This study was supported by Petrobras SA, FAPERJ (Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro), CAPES (Conselho de Aperfeiçoamento de Pessoal de Nível Superior), and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico).
REFERENCES
- Bartnicki-Garcia, S. (1989) The biochemical cytology of chitin and chitosan synthesis in fungi. In: Skjark-Braek, G., Anthonsen, T. and Sandford, P., Eds., Chitin and Chitosan: Sources, Chemistry, Biochemistry, Physical Properties and Applications, Elsevier, Essex, 23-25.
- Deshpande, M.V. (2005) Chitosan in fungi. In: Dutra, P.K., Ed., Chitin and chitosan—Opportunities and Challenges. SSM International Publication, Contai, 59-68.
- Muzzarelli, R. (1994) In vivo biochemical significance of chitin-based medical items. In: Dumitriu, S., Ed., Polymeric Biomaterials, Marcel Dekker, Inc., New York, 179- 197.
- Di Martino, A., Sittinger, M. and Risbud, M.V. (2005) Chitosan: A versatile biopolymer for orthopaedic tissue-engineering. Biomaterials, 26, 5983-5990. doi:10.1016/j.biomaterials.2005.03.016
- Zhao. Y., Park, R. and Muzzarelli, R.A.A. (2010) Chitin deacetylases: Properties and applications. Marine Drugs, 8, 24-46. doi:10.3390/md8010024
- Buranapanitkit, B., Srinilta, V., Ingviga, N., Oungbho, K., Geater, A. and Ovatlarnporn, C. (2004) The efficacy of a hydroxyapatite composite as a biodegradable antibiotic delivery system. Clinical Orthopaedics and Related Research, 424, 244-252. doi:10.1097/01.blo.0000130268.27024.c1
- Chang, K.L., Tsai, G., Lee, J. and Fu, W.R. (1997) Heterogeneous N-deacetylation of chitin in alkaline solution. Carbohydrate Research, 303, 327-332. doi:10.1016/S0008-6215(97)00179-1
- Cantarel, B.L., Coutinho, P.M., Rancurel, C., Bernard, T., Lombard, V. and Henrissat, B. (2009) The carbohydrateactive EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Research, 37, 233-238. doi:10.1093/nar/gkn663
- Okuyasu, K., Kaneko, S., Hayashi, K. and Mori, Y. (1999) Production of a recombinant chitin deacetylase in the culture medium of Escherichia coli cells. FEBS Letters, 458, 23-26. doi:10.1016/S0014-5793(99)01113-8
- Martinou, A., Koutsioulis, D. and Bouriotis, V. (2003) Cloning and expression of a chitin deacetylase gene (CDA2) from Saccharomyces cerevisiae in Escherichia coli. Purification and characterization of the cobalt-dependent recombinant enzyme. Enzyme and Microbial Technology, 32, 757-763. doi:10.1016/S0141-0229(03)00048-6
- Gauthier, C., Clerisse, F., Dommes, J. and Jaspar-Versali, M.F. (2008) Characterization and cloning of chitin deacetylases from Rhizopus circinans. Protein Expression and Purification, 59, 127-137. doi:10.1016/j.pep.2008.01.013
- Christodoulidou, A., Briza, P., Ellinger, A. and Bouriotis, V. (1999) Yeast ascospore wall assembly requires two chitin deacetylase isozymes. FEBS Letters, 460, 275-279. doi:10.1016/S0014-5793(99)01334-4
- Martinou, A., Koutsioulis, D. and Bouriotis, V. (2002) Expression, purification and characterization of a cobaltactivated chitin deacetylase (Cda2p) from Saccharomyces cerevisiae. Protein Expression and Purification, 24, 111- 116. doi:10.1006/prep.2001.1547
- Martinou, A., Bouriotis, V., Stokke, B.T. and Varum K.M. (1998) Mode of action of chitin deacetylase from M. rouxii on partially N-acetylated chitosans. Carbohydrate Research, 311, 71-78. doi:10.1016/S0008-6215(98)00183-9
- Tsigos, I., Martinou, A., Kafetzopoulos, D. and Bouriotis, V. (2000) Chitin deacetylases: New, versatile tools in biotechnology. Trends Biotechnology, 18, 305-312.
- Blair, D.E., Hekmat, O., Schuttelkopf, A.W., Shrestha, B., Tokuyasu, K., Withers, S.G. and Van Aalten, D.M.F. (2006) Structure and mechanism of chitin deacetylase from the fungal pathogen Colletotrichum lindemuthianum. Biochemistry, 45, 9416-9426. doi:10.1021/bi0606694
- Blair, D.E., Schuttelkopf, A.W., Macrae, J.I. and Van Aalten, D.M. (2005) Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor. Proceedings of the National Academy of Science, 102, 15429-10975. doi:10.1073/pnas.0504339102
- Pareek, N., Vivekanand, V., Saroj, S., Sharma, A.K. and Singh, R.P. (2012) Purification and characterization of chitin deacetylase from Penicillium oxalicum SAEM-51. Carbohydrate Polymers, 87, 1091-1097. doi:10.1016/j.carbpol.2011.08.041
- Perrakis, A., Tews, I., Dauter, Z., Oppenheim, A.B., Chet, I., Wilson, K.S. and Vorgias, C.E. (1994) Crystal structure of a bacterial chitinase at 2.3 Å resolutions. Structure, 2, 1169-1180. doi:10.1016/S0969-2126(94)00119-7
- Laemmli, K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T7. Nature, 227, 680-685. doi:10.1038/227680a0
- Gomes, L.P., Oliveira, C.I.R., Da Silva, M.C., Andrade, C.T, Del Aguila, E.M., Silva, J.T. and Paschoalin, V.M.F. (2010) Purificação e Caracterização da Quitinase de Uva (Vitis vinifera L. cv Red Globe) para a produção de quitosana a partir de quitina de camarão. Química Nova, 33, 1882-1886. doi:10.1590/S0100-40422010000900012
- Percot, A., Viton, C. and Domard, A. (2003) Optimization of chitin extraction from shrimp shells. Biomacromolecules, 4, 12-18. doi:10.1021/bm025602k
- Dorresteijn, R., Berwal, L.G., Zomer, G., De Gooijer, C.D., Wieten, G. and Beuvery, E.C. (1996) Determination of amino acids using O-phthalalde-hyde-2-mercaptoeth-anol derivatization. Effect of reaction conditions. Journal of Chromatography A, 724, 159-167. doi:10.1016/0021-9673(95)00927-2
- Brugnerotto, J., Lizardi, J., Goycoolea, F.M., ArgüellesMonal, W., Desbrières, J. and Rinaudo, M. (2001) An infrared investigation in relation with chitin and chitosan characterization. Polymer, 42, 3569-3580. doi:10.1016/S0032-3861(00)00713-8
- Shrestha, B., Blondeau, K., Stevens, W.F. and Hegarat, F.L. (2004) Expression of chitin deacetylase from Colletotrichum lindemuthianum in Pichia pastoris: Purification and characterization. Protein Expression and Purification, 38, 196-204. doi:10.1016/j.pep.2004.08.012
- Cai, J., Yang, J., Du, Y., Fan, L., Qiu, Y., Li, Y. and Kennedy, J. (2006). Purification and characterization of chitin deacetylase from Scopulariopsis brevicaulis. Carbohydrate Polymers, 65, 211-217. doi:10.1016/j.carbpol.2006.01.003
- Saito, Y., Putaux, J-L., Okano, T., Gail, F. and Chanzy, H. (1997) Structural aspects of the swelling of β chitin in HCl and its conversion into a chitin. Macromolecules, 30, 3867-3873.
- Rusu-Balaita, L., Desbrières, J. and Rinaudo, N. (2003) Formation of a biocompatible polyelectrolyte complex: Chitosan-hyaluronan complex stability. Polymer Bulletin 50, 91-98. doi:10.1007/s00289-003-0144-1
- Daly, R. and Hearn, M.T.W. (2005) Expression of heterologous proteins in Pichia pastoris: A useful experimental tool in protein engineering and production. Journal of Molecular Recognition, 18, 119-138. doi:10.1002/jmr.687
- Racusen, D. (1979). Glycoprotein detection in polyacrylamide gel with thymol and sulfuric acid. Analytical Biochemistry, 99, 474-476. doi:10.1016/S0003-2697(79)80035-4
- Tsigos, I. and Bouriotis, V. (1995) Purification and characterization of chitin deacetylase from Colletotrichum lindemuthianum. Journal of Biological Chemistry, 270, 26286-26291. doi:10.1074/jbc.270.44.26286
- Kafetzopoulos, D., Martinou, A. and Bouriotis, V. (1993) Bioconversion of chitin to chitosan: Purification and characterization of chitin deacetylase from Mucor roxii. Proceedings of the National Academy of Sciences of the United States of America, 90, 2564-2568. doi:10.1073/pnas.90.7.2564
- Martinou, A., Kafetzopoulos, D. and Bouriotis, V. (1995) Chitin deacetylation by enzymatic means: Monitoring of deacetylation processes. Carbohydrate Research, 273, 235-242. doi:10.1016/0008-6215(95)00111-6
- Gao, X., Katsumoto, T. and Onodera, K. (1995) Purification and characterization of chitin deacetylase from Absidia coerulea. Journal of Biochemistry (Tokyo), 117, 257- 263. doi:10.1093/jb/117.2.257
- Benson, J.R. and Hare, P.E. (1975) O-phthalaldehyde: Fluorogenic detection of primary amines in the picomole range. Comparison with fluorescamine and ninhydrin. Proceedings of the National Academy of Sciences of the United States of America, 72, 619-622. doi:10.1073/pnas.72.2.619
- Larionova, N.I., Zubaerova, D.K., Guranda, D.T., Pechyonkin, M.A. and Balabushevich, N.G. (2009) Colorimetric assay of chitosan in presence of proteins and polyelectrolytes by using O-phthalaldehyde. Carbohydrate Polymers, 75, 724-727. doi:10.1016/j.carbpol.2008.10.009
- Tokuyasu, K., Ohnishi-Kameyama, M. and Hayashi, K. (1996) Purification and characterization of extracellular chitin deacetylase from Colletotrichum lindemuthianum. Bioscience, Biotechnology, and Biochemistry, 6, 1598- 1603. doi:10.1271/bbb.60.1598
- Win, N.N. and Stevens, W.F. (2001) Shrimp chitin as substrate for fungal chitin deacetylase. Applied Microbiology and Biotechnology, 57, 334-341. doi:10.1007/s002530100741
- Campana-Filho, S.P., De Britto, D., Curti, E., Abreu, F.R., Cardoso, M.B., Battisti, M.V., Sim, P.C., Goy, R.C., Signini, R. and Lavall, R.L. (2007) Extração, estruturas e propriedades de α e β-quitina. Química Nova, 30, 644- 650. doi:10.1590/S0100-40422007000300026
- Aam, B.B., Heggset, E.B., Norberg, A.L., Sorlie, M., Varum, K.M. and Eijsink, V.G.H. (2010) Production of chitooligosaccharides and their potential applications in medicine. Marine Drugs, 8, 1482-1517. doi:10.3390/md8051482
- Yalpani, M. and Pantaleone, D. (1994) An examination of the unusual susceptibility of aminoglycans to enzymatic hydrolysis. Carbohydrate Research, 256, 159-175. doi:10.1016/0008-6215(94)84235-3

