American Journal of Molecular Biology
Vol.2 No.3(2012), Article ID:20983,8 pages DOI:10.4236/ajmb.2012.23023
Expression and subcellular localization of a novel gene Bm-X of the silkworm, Bombyx mori
![]()
Institute of Biochemistry, Zhejiang Sci-Tech University, Hangzhou, China
Email: *yaozhou@chinagene.com
Received 7 March 2012; revised 6 April 2012; accepted 19 April 2012
Keywords: Bombyx mori; Novel Gene; Expression; Subcellular Localization; Bm-X
ABSTRACT
Based on the large-scale sequencing of cDNA library from silkworm pupae, the cDNA of a previously unknown novel gene with blank research background was identified and temporarily named Bm-X. The length of this cDNA is 778 bp. We obtained its ORF for further study by bioinformatics analysis. It is 444 bp and encodes a protein of 147 amino acid residues, with a predicted molecular weight (MW) of 16.4 kD and an isoelectric point (pI) of 3.69. In this study, we successfully constructed a recombinant plasmid pET- 28a(+)-Bm-X and expressed it in Escherichia coli. We expressed the fusion protein rBm-X which purified by Ni-affinity chromatography and used it to produce polyclonal antibodies against Bm-X for Western blot analysis. Our analysis revealed that Bm-X was expressed in the larval midgut, the epidermis and the silk gland. In addition, the subcellular localization analysis of silkworm ovary epithelial cells (BmN cells) showed that Bm-X protein was located both in cytoplasm and nucleus, with higher localization signal in the cytoplasm than in nucleus. Our findings indicate that Bm-X gene is a novel species-specificity gene and its expression product can be detected in tissues of the fifth silkworm instar larvae and BmN cells.
1. INTRODUCTION
Silkworm (Bombyx mori) is a holometabolic insect which has complete biology background and is a model organism of lepidoptera insects. Researchers study the silkworm intensively to excavate its potential economic value and explore the molecular mechanism of the physiological development in lepidoptera insects [1].
The silkworm genome consists of 28 pairs of chromosomes containing about 4.8 billion base pairs. After complete sequence determination and analysis of silkworm genome, more than 160,000 complete genes and 7000 gene fragments were obtained. Silkworm has more than 20,000 genes and over 25% of them are novel genes [2,3]. Recently, we have constructed a silkworm pupae cDNA library from which isolated 2400 cDNA sequences were isolated. Our library contains many novel genes of unknown function [4]. In our efforts to clarify these unknown genes, we have previously reported the selective expression of the silkworm troponin C gene during silkworm development [5]. Wang demonstrated that the expression of BmCRABP (silkworm Cellular retinoic acid binding protein) gene had effect on the physiological function of atRA [6].
We have previously characterized novel genes in the silkworm using sequence homology comparison and recombinant expression studies. We have demonstrated successful studies, such as the cloning, expression and localization study of a novel Sulfotransferase gene from Bombyx mori [7], the determination of a novel lebocinlike gene from eri-silkworm [8], a novel cuticle protein gene BmCPG1 in the silkworm with Ecdysteroid-dependent expression [9], and a Bombyx mori gene Bmchi-h encoding a protein was found which is homologous to bacterial and baculovirus chitinases [10,11]. So far, there have been few studies of the silkworm novel genes which lack previous understanding and have low homology with any known genes. Thus, there is a need to find an appropriate method to identify the localization, expression and function of these unknown genes in Bombyx mori. This is the main objective of this study.
In this study, we cloned a novel gene Bm-X of silkworm from a silkworm pupae cDNA library constructed. We successfully constructed a recombinant plasmid pET- 28a(+)-Bm-X and expressed it in Escherichia coli (E. coli). The recombinant rBm-X protein was purified Niaffinity chromatography. Immunization of rabbit with the recombinant protein generated high titer (1:8000) polyclonal antibodies, measured by ELISA. Western blotting analysis showed that the antibody could bind rBm-X specifically. Subsequently, subcellular localization analysis of Bm-X gene expression in the fifth instar larvae of silkworm was performed by using these polyclonal antibodies. Our work laid a good foundation for further functional study of the novel gene Bm-X.
2. MATERIALS AND METHODS
2.1. Bacterial Strain
E. coli strains TG1 and Rosseta (DE3) were grown at 37˚C in LB medium, pH 7.5, containing 5 g yeast extract, 10 g tryptone peptone, and 10 g NaCl per liter. Recombinant E. coli strains were grown in LB medium with kanamycin (50 μg/mL) and chloromycin (34 μg/mL).
2.2. Insects and Tissues Preparation
The Qingsong × Haoyue strain of Bombyx mori were reared on fresh mulberry leaves under standard conditions. We isolated the silk gland, head, midgut, epidermis, genital, malpighian tubule, stigma, and fatty body from day 4 and day 5 larvae of the 5th instar. These tissues were washed several times with ice-cold phosphate-buffered saline (PBS, pH 7.4), weighed, and then homogenized in 5 volumes of protein extraction buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 5 mM EDTA, 1 mM dithiothreitole, 0.5% NP-40, 0.5% sodium deoxycholate) containing protease inhibitor (100 μg/mL phenylmethane sulfonyl-fluoride, 5 μg/mL Aprotinin). Homogenates were centrifuged at 15,000 g for 15 min, and the supernatant was collected for Western blot analysis. Protein concentrations were determined by the Bradford method; bovine serum albumin (BSA; Sigma) was used as a standard [12].
2.3. Cloning of Bm-X and Construction of the Recombinant Plasmid
Using our in-house plasmid, TG1-pHelix-Bm-X, as a template, we performed the polymerase chain reaction (PCR) to amplify the Bm-X open reading frame (ORF). We designed a primer pair based on the Bm-X cDNA sequence and introduced restriction endonuclease recognition sites for BamH I and Xho I at the 5’-ends of the primers for subcloning purpose. The sense primer was 5’-CCCGGATCCATGCCGCTGAACAAAACAAC-3’and the antisense primer was 5’-CCGCTCGAGTCAT GACTCGGTTTTATTGTC-3’. The PCR program consisted of a preliminary denaturation step at 94˚C for 5 min, followed by 30 cycles of amplification (denaturation at 94˚C for 30 s, annealing at 55˚C for 30 s, and extension at 72˚C for 1 min), and a final extension at 72˚C for 10 min. PCR products were digested with BamH I and Xho I (Promega), separated by electrophoresis on a 1% agarose gel, and purified by the PCR Rapid Purification Kit (BioDev-Tech). Purified PCR products were subcloned into the expression vector pET-28a (Amersham Biosciences) by T4 DNA ligase (Promega). The expression plasmids containing the PCR products inserted were transformed into competent E. coli (TG1 strain), then picked white colonies. We identified positive colonies that contained the recombinant plasmid among white colonies. Then recombinant plasmids were extracted and digested with BamH I and Xho I, followed by separation and analysis on 1% agarose gel electrophoresis.
2.4. Sequence Analysis
Using the BLAST search of Genbank [13] (http://www.ncbi.nlm.nih.gov/BLAST/), we did not find any sequence similarities with the Bm-X sequence. We determined the theoretical molecular weight and isoelectric point by Peptide Mass [14] (http://us.expasy.org/tools/peptide-mass.html). We also performed secondary structure prediction (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_gor4.html), hydrophobicity prediction (http://www.expasy.org/cgi-bin/protscale.pl), and signal peptide prediction (http://www.cbs.dtu.dk/services/SignalP/) by on-line biological information tools.
2.5. Expression and Purification of rBm-X
The recombinant plasmid pET-28a-Bm-X was transformed into competent E. coli cells [Rosseta (DE3) strain] and the expression of the His-tagged fusion protein with His-tag was induced with 1 mM isopropyl thiogalactoside for 4 h [15]. Recombinant Bm-X (rBm-X) was purified by HiTrap Chelating HP (Amersham), according to the manufacturer’s instructions. The eluted rBm-X was freeze-dried in an alpha 2 - 4 Freeze Dryer (Christ) and then stored at –20˚C.
2.6. Preparation and Purification of Anti-Bm-X Serum
We injected intraperitoneally a New Zealand White rabbit with a mixture of 1mg purified recombinant Bm-X and Freund’s complete adjuvant (Dingguo). The rabbit was boosted three times with 0.5 mg recombinant Bm-X in Freund’s incomplete adjuvant in one week intervals. Serum was derived from blood collected 10 days after the third injection and purified by HiTrap Protein A HP (Amersham) according to the manufacture’s instructions. After the IgG was filtered through Ultracel PLCHK (Millipore), the purified anti-Bm-X IgG was dissolved in 50% glycerol and stored at –80˚C.
 (a)
(a) (b)
(b)
Figure 1. (a) ORF sequence and deduced amino acid sequence of Bm-X; (b) Gene sequence analysis of Bm-X ORF. The BLAST analysis of the ORF of Bm-X, each ORF from other three ATGs to terminator (underlined in Figure 1(a)) has no similarity to other species.
2.7. SDS-PAGE and Immunoblot Analysis
Equal quantities of proteins from various silkworm tissues were separated by SDS-PAGE under reducing conditions, as described by Laemmli (1970). The proteins separated by electrophoresis were then transferred electrophoretically onto polyvinylidene difluoride membranes. Following incubation with purified anti-Bm-X IgG, membranes were washed and incubated with goat antirabbit IgG antibody conjugated to Alexa Fluor 680 (Molecular probe). Finally, membranes were washed three times with Tris-buffered saline plus Triton X-100, twice with double-distilled H2O, and subsequently scanned by the Odyssey Infrared Imaging System (LI-COR) at 700 nm.
2.8. Subcellular Localization
BmN cells were seeded arbitrarily in the dish (Bio-line Instruments) which was used specially for Confocal Microscope. After 12 h, culture medium was removed, and cells were rinsed twice with 1 mL phosphate-buffered saline (PBS), fixed in 3.7% formaldehyde at 25˚C for 10 min. Then the cells were blocked with 3% BSA at 37˚C for 2 h. After that, the cells were incubated with anti-BmX IgG (dilution 1/200) at 4˚C for 12 h (the cells incubated with serum as negative control). After washing three times in PBST (PBS + 0.05% Tween-20, 10 min each), cells were incubated with goat anti-rabbit antibody (dilution, 1/2000, Cy3 labeled, Promega) and 4’-6- Diamidino-2-phenylindole (dilution, 1/2000) at 37˚C for 2 h. Following three times washing with PBST (10 min each), cells were analyzed by Nikon ECLIPSE TE2000- E Confocal Microscope with image analysis software EZ-C1.
3. RESULTS
3.1. Cloning and Bioinformatics Analysis of Bm-X
Using specific PCR primers on our in-house plasmid TG1-pHelix-Bm-X, we amplified the cDNA of the B. mori-X gene (Bm-X). We have sequenced the Bm-X cDNA and submitted the sequence to GenBank (data on accessing). Sequence analysis revealed that the ORF of Bm-X is 444 bp, and encoded a putative protein containing 147 amino acids. We predicted that the molecular mass of the putative protein was 16.4 kDa (Figure 1(a)). The nucleotide sequence of Bm-X and the deduced amino acid sequence of the protein are shown in Figure 1(a). We also used BLAST to analyse the full ORF of Bm-X and each of the three shorter ORFs starting from three other ATG codons. The result showed that there was no similarity to other species (Figure 1(b)). This indicated that the Bm-X gene is a species-specific gene of Bombyx mori without any previously known homologues.
3.2. Characteristics of Deduced Bm-X Protein by Bioinformatics Analysis
BLASTX analysis of the translation product deduced from the ORF showed that Bm-X has no significant homology with other proteins in the GenBank database. We conclude that Bm-X is likely a novel species-specific gene and Bm-X protein is important to silkworm. Bioinformatics prediction showed that Bm-X protein contains 12 functional sites (Figure 2), carried with no signal peptide, and has a seconddary structure chiefly consisting of alpha helix and random coil (Figure 3).
3.3. Expression and Purification of Recombinant Bm-X
The Bm-X was inserted into the expression vector pET-28a with a BamH I site to construct a recombinant plasmid named pET-28a-Bm-X. The recombinant plasmid pET- 28a-Bm-X was then transformed into competent E. coli cells. We confirmed the expression of recombinant Bm-X in E. coli by SDS-PAGE (Figure 4). We estimated that the molecular weight of recombinant Bm-X was 19.96 kDa, which included a 3.56 kDa His-tag. We immunized New Zealand White rabbits with the purified recombinant protein to obtain anti-Bm-X rabbit serum. Using the anti-Bm-X rabbit serum, we detected a strong signal of 19.96 kDa corresponding to recombinant Bm-X in induced E. coli lysate, whereas no signal was detected in the non-induced E. coli lysate (Figure 5).
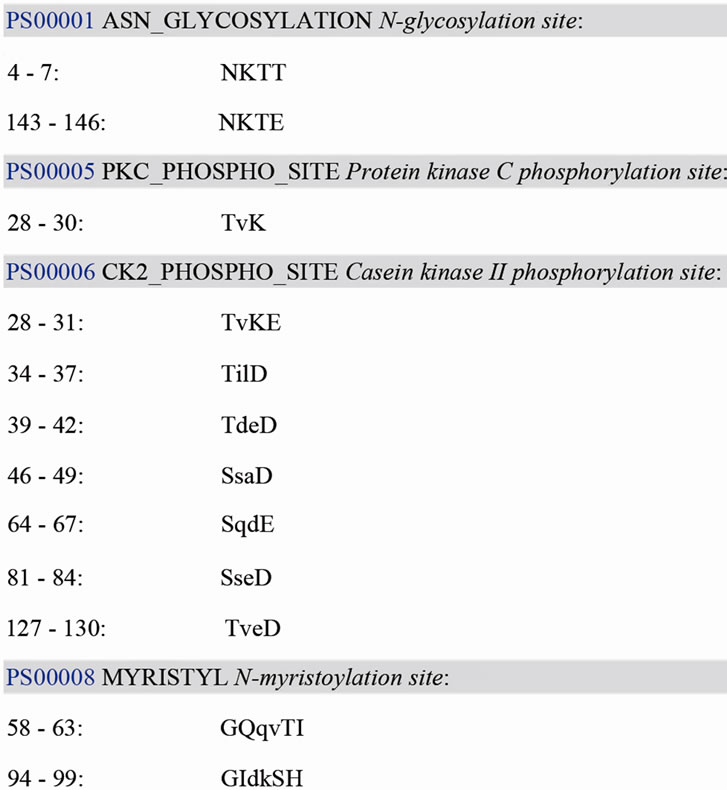
Figure 2. The prediction of Bm-X functional sites.
 (a)
(a) (b)
(b) (c)
(c)
Figure 3. Characteristics of deduced Bm-X protein by bioinformatics analysis. (a) Signal peptide prediction of silkworm Bm-X protein; (b) Hydrophobicity prediction of silkworm Bm-X protein; (c) Secondary structure prediction of silkworm Bm-X protein.
3.4. Tissue Distribution of Bm-X Protein
In order to detect the expression level of Bm-X protein in each tissue, we extracted proteins from the head, midgut, genital, silk gland, epidermis, fatty body, stigma, and Malpighian tubule of the fifth instar larva, and used Western blot analysis to determine the level of Bm-X in each tissue. Immunoblots of these protein extracts re-

Figure 4. Purification of protein fractions obtained from E. coli Rosseta (DE3) expressing the recombinant Bm-X. Samples were resolved by 12% SDSpolyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions (lane 1: lysate from E. coli without induction; lane 2: lysate from E. coli after induction; lane 3: lysate from purified recombinant Bm-X; lane M: size markers).

Figure 5. Western blot analysis of recombinant Bm-X induction. Samples were resolved by 12% SDS-PAGE under reducing conditions (lane 1: lysate from purified recombinant Bm-X; lane 2: lysate from E. coli without induction; lane M: size markers; lane 3: Western blot result of purified rBm-X; lane 4: Western blot result of Rosseta (DE3)-pET-28a-Bm-X without induction).
vealed that anti-Bm-X serum reacted with a protein over 20.1 kDa in extracts isolated from the midgut, epidermis and silk gland, but did not react with any protein in extracts isolated from other tissues (Figure 6). The amount of the Bm-X protein was the highest in extracts from the epidermis and lower in extracts from the midgut and silk gland.
3.5. Subcellular Localization of Bm-X Protein
We used the BmN cells to observe the subcellular localization of endogenous Bm-X protein. Immunostaining showed that Bm-X protein localized in both the cytoplasm and nucleus. But the fluorescence localization signals were stronger in cytoplasm than in nucleus, even in the dividing cells (Figure 7).
4. DISCUSSION
We described the cloning, expression, and subcellular localization of a novel gene Bm-X in the study. We have also obtained and purified an anti-Bm-X serum for further studies. As far as we know, our laboratory is the first to report tissue-specific expression and subcellular localization of this species-specific gene in Bombyx mori.
By bioinformatics tools, we confirmed a novel gene Bm-X of Bombyx mori from our silkworm pupae cDNA library. Bm-X gene belongs to silkworm genome, but lacks any significant homology with other species. It is a species-specific gene of Bombyx mori without any previous knowledge. We predicted the molecular characteristics of Bm-X protein by bioinformatics. Bioinformatics analysis can be informative for the design of experiments to study unknown genes, for example the predicted hydrophilicity was beneficial to protein purification and predicted functional sites were useful for functional studies.
In this study, we obtained polyclonal antibodies specific for the Bm-X protein. In order to purify the natural Bm-X from the fifth instar larvae, we extracted total proteins of this larval instar and salted out with 100% saturated ammonium sulfate. These proteins were then used in the Western blot analysis. The results showed that the antibody has high degree of specificity with Bm-X protein. The Western blot analysis revealed that a protein band over 19.96-kDa appeared in the silkworm protein extracts (Figure 4), indicating that the molecular weight of endogenous Bm-X was approximate 20 kDa. However, there was a significant difference between the molecular weight of the endogenous Bm-X protein and that expressed from bacteria. We believe that the reason for such a difference lies in post-translational modification of natural Bm-X protein. This modification may be related to the two predicted glycosylation sites of natural

Figure 6. Expression level of Bm-X in different tissues of 5th instar larva of Bombyx mori by Western blot analysis. Protein extracts from the head (H), midgut (Md), Genital (G), silk gland (S), epidermis (E), fatty body (F), stigma (T), and Malpighian tubule (M) of 5th instar larva were subjected to immunoblot analysis. Here we use rBm-X as a positive control.

Figure 7. The subcellular localization of Bm-X protein. 1: Visible light images; 2: Cy3 fluorescence images; 3: DAPI fluorescence images; 4: Mix images control. Negative control; A: Positive result by 40× amplified; B: Positive result by 40 × 2.5 amplified.
Bm-X protein.
During the fifth instar larva, the expression of Bm-X is distributed broadly in some tissues, including the silk gland, midgut and epidermis as confirmed by our Western blot analysis. The tissue distributions differed greatly during pupation of Bombyx mori. In addition, we did not detect obvious signals of Bm-X protein in the pupae, suggesting that Bm-X may play a role in molecular mechanisms in Bombyx mori larvae pupation. Our results were reproducible in repeated experiments, and we observed that the sensitivity of detection from fluorescence western blot was 20 times higher than western blot with DAB dried, suggesting that the expression level of Bm-X in these three tissues may be low.
The subcellular localization analysis of Bm-X protein showed that Bm-X protein localizes in both the cytoplasm and nucleus. The fluorescence localization signals were stronger in cytoplasm than in nucleus, even there is no change in the dividing cells. The scanning laser tomography also showed that Bm-X protein is uniformly distributed in BmN cells.
We have identified a novel Bm-X gene which is is specific to Bombyx mori. We immunized New Zealand rabbits with the recombinant Bm-X to obtain the antiBm-X polyclonal antibodies which showed high degree of specificity against rBm-X. We have found that Bm-X is expressed in the epidermis, silk gland, and midgut of the silkworm. However, the specific function of Bm-X in various periods of development in silkworm requires further research. One possible approach to determine these functions would be RNA interference with dsRNA designed to inhibit the expression of Bm-X.
5. ACKNOWLEDGEMENTS
This work was supported by financial grants from the National High Technology Research and Development Program (No. 2011AA100603), Zhejiang Natural Science Foundation (No. Y3090304, Y3090339, Y31- 10051, Y3110354) and Zhejiang Educational Foundation (Y200909740, Y201019098).
REFERENCES
- Zhang, Y., Huanga, J.H., Jia, S.H., et al. (2007) SAGE tag based cDNA microarray analysis during larval to pupal development and isolation of novel cDNAs in Bombyx mori. Genomics, 90, 372-379. doi:10.1016/j.ygeno.2007.05.005
- Huang, N., Clem, R.J. and Rohrmann, G.F. (2011) Characterization of cDNAs encoding p53 of Bombyx mori and Spodoptera frugiperda. Insect Biochemistry and Molecular Biology, 41, 613-619. doi:10.1016/j.ibmb.2011.03.014
- Xia, Q.Y., Zhou, Z.Y., Lu, C., et a1. (2004) A draft sequence for the genome of the domesticated silkworm Bombyx mori. Science, 306, 1937-1940. doi:10.1126/science.1102210
- Zhang, Y.Z., Chen, J., Nie, Z.M., et al. (2007) Expression of open reading frames in silkworm pupal cDNA library. Applied Biochemistry and Biotechnology, 136, 327-343. doi:10.1007/s12010-007-9029-3
- Chen, J.Q., Chen, J., Gai, Q.J., et al. (2007) Molecular characterization and immunohistochemical localization of a novel troponin C duiing silkworm development. Cell and Tissue Research, 331, 725-738. doi:10.1007/s00441-007-0516-1
- Wang, X.J., Chen, J., Lv, Z.B., et al. (2007) Expression and functional analysis of the cellular retinoic acid binding protein from silkworm pupae (Bombyx mori). Journal of Cellular Biochemistry, 102, 970-979. doi:10.1002/jcb.21333
- Hattori, K., Hirayama, M., Suzuki, H., et al. (2007) Cloning and expression of a novel sulfotransferase with unique substrate specificity from Bombyx mori. Bioscience, Biotechnology, and Biochemistry, 71, 1044-1051. doi:10.1271/bbb.60703
- Bao, Y.Y., Yamano, Y. and Morishima, I. (2005) A novel lebocin-like gene from eri-silkworm, Samia Cynthia ricini, that does not encode the antibacterial peptide lebocin. Comparative Biochemistry and Physiology, 140, 127- 131. doi:10.1016/j.cbpc.2004.09.022
- Suzuki, Y., Matsuoka, T., Iimura, Y., et al. (2002) Ecdysteroid-depedent expression of a novel cuticle protein gene BmCPG1 in the silkworm, Bombyx mori. Insect Biochemistry and molecular Biology, 32, 599-607. doi:10.1016/S0965-1748(01)00136-9
- Daimon, T., Hamada, K., Mita, K., et al. (2003) A Bombyx mori gene, Bmchi-h, encodes a protein homologous to bacterial and baculovirus chitinases. Insect Biochemistry and Molecular Biology, 33, 749-759. doi:10.1016/S0965-1748(03)00084-5
- Zhang, H.B., Liu, M.Y., Tian, Y.J., et al. (2011) Comparative characterization of chitinases from silkworm (Bombyx mori) and bollworm (Helicoverpa armigera). Cell Biochemistry and Biophysics, 61, 267-275. doi:10.1007/s12013-011-9196-2
- Bradford, M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248-254. doi:10.1016/0003-2697(76)90527-3
- Altschul, S.F., Madden, T.L., Schaffer, A.A., et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research, 25, 3389-3402. doi:10.1093/nar/25.17.3389
- Wilkins, M.R., Lindskog, I., Gasteiger, E., et al. (1997) Detailed peptide characterization using PEPTIDEMASSa world-wide-web-accessible tool. Electrophoresis, 18, 403-408. doi:10.1002/elps.1150180314
- Joseph, S. and David, W.R. (2002) Molecular cloning: a laboratory manual. CSHL Press, New York.
NOTES
*Corresponding author.

