Advances in Bioscience and Biotechnology
Vol.4 No.2A(2013), Article ID:28440,5 pages DOI:10.4236/abb.2013.42A038
The Central Cattle Breeding and Dairy Farm, Bangladesh waste contributes in emergence and spread of aminoglycoside-resistant bacteria
![]()
1Department of Biochemistry and Molecular Biology, Jahangirnagar University, Dhaka, Bangladesh
2Central Cattle Breeding and Dairy Farm (CCBDF), Dhaka, Bangladesh
3Department of Microbiology, University of Dhaka, Dhaka, Bangladesh
Email: *drsahmed_bmbju@yahoo.com
Received 21 December 2012; revised 22 January 2013; accepted 30 January 2013
Keywords: Aminoglycosides; Resistant Bacteria; Cattle Diseases; Veterinary Clinical Healthcare Waste
ABSTRACT
Aminoglycosides are one of the categories of antibiotics most frequently used in treating several cattle diseases at the Central Cattle Breeding and Dairy Farm (CCBDF), Savar, Dhaka, Bangladesh. Untreated veterinary clinical healthcare waste (VCHW) of diseased cattle at CCBDF which directly disposed to surrounding may contribute to the antibiotic resistant bacteria pollution (ARB) pollution. The investigation analyses the role of VCHW of CCBDF in spreading ARB. Here we studied:1) veterinary clinical data and antibiotics treatment history; 2) total and resistant bacteria counts in fecal samples of healthy and diseased cattles as well as VCHW of CCBDF; and 3) finally, data analysis to estimate the burden of VCHW of CCBDF in the pollution of environment with aminoglycoside antibiotics resistant bacteria. The results conclusively demonstrate the spread of 3 different aminoglycoside antibiotics, namely gentamycin, kanamycin and streptomycin resistant bacteria in the surrounding environment alarmingly with high significant value (p < 0.01 - 0.05). This study reveals the risks to the cattle as well as public health posed by the random VCHW disposal at the CCBDF, Bangladesh.
1. INTRODUCTION
Selection of antibiotic resistant bacteria (ARB) occurred due to the extensive use of antibiotics in healthcare as medicine and prophylaxis, and also as growth promoter in animal feeds [1,2]. Over or irrational use of antibiotics further exaggerated the situations and has led to selection of multi-drug resistant bacteria in humans and cattle, respectively [3,4]. Members of the Enterobacteriaceae family of bacteria are medically important as infectious agents, exhibit antibiotic resistance and are present in large numbers in the animal gut [5,6]. Enteric bacteria such as Escherichia coli, Enterococcus faecalis, and Salmonella spp. became resistant to multiple antibiotics given to animals and VCHW pollute environment with these resistant bacteria as well as with un-metabolized antibiotics or its active metabolites; and thus made available to humans and farm animals [7,8]. Furthermore, these enteric bacteria are found in the intestinal tract of humans and cattle, providing a potential reservoir for antimicrobial gene pools and may cause for zoonoses and transfer of resistant-gene transversely and horizontally [9-11]. The situation of antimicrobial resistance become alarming worldwide and resistance to antibiotics is rapidly outpacing our ability to synthesize new drugs. Hence, emerging of ARB becomes a serious growing problem in modern medicine. In addition to human setting, antibiotics are extensively used in veterinary medicine for the treatment of infections, growth enhancement, and prophylaxis in animal feeds. Knowledge about relative composition of antibiotics resistant-bacteria in nonnosocomal communities is limited. Lack of surveillance data is especially evident in important agricultural settings in Bangladesh, such as cattle dairy farm waste disposal on the natural environment. Antibiotics are not metabolized completely in the body and may be expelled out mainly through urine and feces. Thus waste water carries both the resistant bacteria and un-metabolized antibiotics. Un-treated liquid and solid wastes discharged directly to sewerage systems may further worsen the situations. It remains unclear to what extent dairy farm environment harbors multi-drug resistant bacteria. Once the surveillance data are known on the emergence of resistant bacterial population in an agricultural setting like cattle dairy farm, it would be possible to invoke measures and environmental management system to reduce the conditions that lead to antibiotic resistance, and thus limit conditions that foster the spread or fixation of resistant infectious bacteria in the dairy farm environments. The aim of this investigation is to assess the burden of untreated clinical healthcare waste (CHW) of the Central Cattle Breeding and Dairy Farm (CCBDF), Savar, Dhaka, Bangladesh which directly disposed to surrounding in spreading of ARB; and demonstrated that CHW of CCBDF pollute significantly it’s surrounding with aminoglycoside ARB.
2. MATERIALS AND METHODS
2.1. Cattle Diseases and Antibiotic Use at the CCBDF
Data of veterinary healthcare and antibiotics used at the CCBDF during the study periods were collected from CCBDF records.
2.2. Sample Collection
Fecal as well as waste-water samples (WWSs) were collected. Fecal samples were collected from healthy as well as diseased cattle. During the sampling period, cattle were suffering from mastitis and pneumonia. Samples were collected from cattle suffering either from mastitis or pneumonia. Samples were collected aseptically in sterile tubes by maintaining standard sanitary and clinical safety measures under supervision of a trained veterinary technician of CCBDF; and immediately transported to Laboratory at the Department of Biochemistry and Molecular Biology, Jahangirnagar University, Savar, Dhaka. About 15 g of feces and 15 ml of waste-water were collected for each sample. Samples were collected in triplicate from healthy cattle (n = 3) and from diseased cattle (mastitis, n = 3; pneumonia, n = 3). Fecal samples were collected three times on the same day either from each healthy or diseased cattle. In case of the WWSs, samples were collected thrice from each location. WWSs were collected from 10 different locations of the sewerage lines connected to the healthy as well as to the recovery sheds of CCBDF. WWSs were the mixture of cattle feces and urine, washed away from the cattle sheds with the flushing of water and drained into a the common sewerage line. In total, 57 samples (fecal samples, n = 27 and WWSs, n = 30) were collected from CCBDF.
2.3. Sample Processing
After the samples were transferred to the research laboratory, the fecal samples were immediately processed. In the processing procedure, at first 10 g of fresh feces were diluted with 10 mL of sterile saline (0.8%). The diluted fecal samples were first passed through sterile gauze and then filtrated through sterile filter paper. The filtrate was collected and used for further research. In order to enumeration of bacteria, samples were serially diluted with sterile saline (0.8%) up to 0 to 10−6. In case of wastewater samples, 10 mL of the sample was directly filtered through sterile filter paper. Then the filtrate was collected and used for enumeration of bacteria by dilution of the samples with sterile saline (0.8%) up to 0 to 10−6.
2.4. Preparation of Aminoglycosides and Supplementation to Bacteriological Media
For isolation of the ARB, the Plate Count Agar (PCA) was individually supplemented either with gentamycin or kanamycin or streptomycin as per the Clinical and Laboratory Standards Institute (CLSI) approved standard [12]. Stock solutions of aminoglyside antibiotics dissolved in double distilled water was individually prepared, sterilized through filtration with 0.20 µm disposable syringe filter (Sartorius, Germany) under a cleaned laminar air flow cabinet (ESCO, Singapore) and supplemented to the media as shown below (Table 1).
2.5. Preparation of Plate Count Agar (PCA) Media and Enumeration of Total Bacterial Count
PCA was made according to the instruction as described in the container of the media. The agar media were sterilized at 121˚C temperature and 15 lb pressure for 15 minutes. The media were then cooled at 45˚C and maintained in a molten condition at this temperature until pouring onto the plate. 0.1 mL of the samples from different dilutions (102, 103, 104, 105 and 106) was poured into the petri-dishes with sterile pipettes. The samples were mixed well with the media by a spreader. The plates were then placed within an incubator (SANYO, MIR-162, Japan) in an inverted position at 37˚C temperature for overnight. After overnight growth, plates with 25 to 250 bacterial colonies were counted [13]. The total bacterial count (TBC) was calculated by multiply ing the count with dilution factor. The TBC per ml were calculated by number of bacteria × dilution factor. .
Table 1. Stock solutions of aminoglycosides and supplementation.

2.6. Isolation of Resistant Bacteria and Preservations
Single colonies were randomly picked from PCA plates and transfer to nutrient agar (NA) media containing the three aminoglycoside antibiotics (gentamycin, kanamycin, streptomycin) for further characterizations. ARBs were preserved in presence of appropriate antibiotics on NA plate for routine work and 15% glycerol broth were used for long-time preservation at −20˚C.
2.7. Statistical Analysis
All data were presented as mean ± Standard Error Mean, SEM. Means values were compared by independent sample t-test using the statistical program “SPSS 12.0 for Windows”.
3. RESULTS AND DISCUSSION
In the present study, we have assessed the impact of VCHW of CCBDF on the possible emergence and spread of ARB in the natural environment. Common antibiotics used in treating different cattle diseases at CCBDF are shown in Table 2.
The most frequently used antibiotics were found to be aminoglycosides such as gentamycin, kanamycin and streptomycin. Among the aminoglycosides, streptomycin was frequently used for treating tuberculosis (TB). Specifically, gentamycin and kanamycin were used for treating mastitis and pneumonia, respectively (records not shown).
TBC in fecal samples from healthy and diseased cattle as well as in waste water samples (WWSs) of CCBDF are shown in Table 3. We found that TBC/mL were
Table 2. Antibiotics used in treating common cattle diseases at the CCBDF.

Table 3. Total bacterial count in fecal samples: healthy cattle vs., diseased cattle.
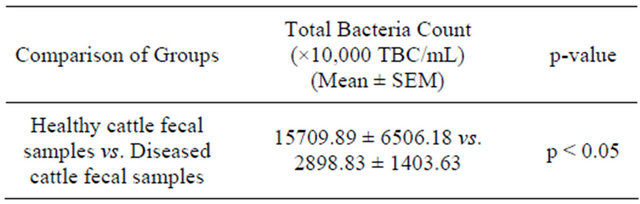
significantly higher (p < 0.05) in the fecal samples of the healthy cattle than that of the diseased cattle (Table 3).
Results presented as mean ± Standard Error Mean, SEM (fecal samples of healthy cattle, n = 3 and fecal samples of diseased cattle, n = 6). Means (fecal samples of healthy cattle vs. fecal samples of diseased cattle) were compared by independent sample t-test using the statistical program “SPSS 12.0 for Windows”.
Similarly, TBC/mL were found significantly higher in fecal diseased cattle samples compared to WWSs (p < 0.01) (Table 4).
Results presented as mean ± Standard Error Mean, SEM (fecal samples of diseased cattle, n = 6 and wastewater samples from CCBDF sewerage lines, n = 11). Means (fecal samples of diseased cattle vs. and wastewater samples from CCBDF sewerage lines) were compared by independent sample t-test using the statistical program “SPSS 12.0 for Windows”.
Next to determine the percentage of ARB resistant to gentamycin or kanamycin or streptomycin in different samples mentioned above by PCA method supplemented with either gentamycin or kanamycin or streptomycin (p < 0.01). When we compared the percentage of ARB to different aminoglycosides examined in fecal samples of healthy cattle to that of diseased cattle, we did not find any significant differences (Table 5).
Results represented as mean ± SEM (healthy cattle fecal samples, n = 3 vs. diseased cattle fecal samples, n = 6). Means (healthy cattle fecal samples vs. diseased cattle fecal samples) were compared by doing independent sample t-test using the statistical program “SPSS 12.0 for Windows”.
However, when we compared the % of ARB to different aminoglycosides (gentamycin or kanamycin or strepto-mycin) in fecal samples of diseased cattle to that of waste-water samples, significant differences were observed in case of kanamycin (p < 0.01) and streptomycin (p < 0.05) (Table 6).
Results are represented as mean ± SEM (Diseased cattle fecal samples, n = 6 vs. waste-water samples from sewerage lines, n = 11). Means (Diseased cattle fecal samples vs. waste-water samples from CCBDF sewerage lines) were compared by doing independent sample t-test using the statistical program “SPSS 12.0 for Windows”.
The impact of streptomycin, the most frequently used antibiotic to treat cattle tuberculosis at CCBDF on the
Table 4. Total bacterial count: fecal samples of diseased cattle vs., waste-water samples from CCBDF sewerage lines.

Table 5. Percentage of antibiotic resistant bacteria resistant to aminoglycoside: healthy cattle fecal samples vs. diseased cattle fecal samples.
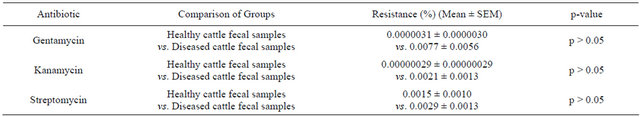
Table 6. Percent of resistant bacteria to aminoglycoside antibiotics: diseased cattle fecal samples vs. waste-water samples from sewerage lines.

possible emergence of streptomycin resistant bacteria in the natural environment of CCBDF was also assessed, we compared the % of resistant bacteria in waste-water samples collected from different locations of CCBDF sewerage lines. We found that the highest percentage (%) of bacteria were resistant to streptomycin in almost all the locations (data not shown).
Enumeration of the TBC/mL grown on PCA media revealed that the fresh fecal samples collected from healthy cattle contained significantly (p < 0.05) higher numbers of colony forming units of bacteria than those collected from diseased cattle (Table 3). It was likely as the normal intestinal flora of healthy cattle did not come under any antibiotic pressure. On the other hand, treatment of diseased cattle with antibiotics destroyed the pathogenic microorganisms as well as the normal intestinal microbial flora. Moreover, it was possible that growth of the normal intestinal flora in the diseased cattle were inhibited by the overgrowth of various pathogenic microorganisms resulting in the overall reduction of TBC.
We also found that the TBC were significantly (p < 0.01) higher in the fecal samples from diseased cattle than those in the waste-water samples from the CCBDF sewerage lines (Table 4). This difference might be due to the dilution of fecal wastes with the flushing of water used to clean the sheds of the cattle. It was also possible that a portion of the antibiotics administered into the diseased cattle were expelled out with the feces in unmetabolized active form and these have exerted bactericidal effect on the natural bacterial flora in the environment.
Enumeration and comparison of the TBC among the different categories of samples revealed that the % of resistant bacteria to kanamycin or streptomycin in fecal samples of diseased cattle were significantly (p < 0.01 for kanamycin and p < 0.05 for streptomycin) lower than those in waste-water samples (Table 6). We assume that these significant differences were due to the frequent use of these two aminoglycoside antibiotics in various cattle diseases by the veterinary physicians, which in turn were expelled with the feces and urine in part in un-metabolized active form and led to the emergence of overall bacteria resistance in the natural environment of CCBDF.
The lower percent of resistant bacteria in the diseased cattle fecal samples than the waste-water samples indicates the generation of antibiotic resistance in environment rather than inside the body i.e. in the gut. It was also possible that a fraction of the bacterial population developed resistance in the gut of the diseased cattle, while being treated with antibiotics and the untreated VCHW when discharge to the surroundings pollutes environment with ARB. After their disposal to the environment, these resistant bacteria might play an important role as resistant gene pool pollution and spreading the resistant markers through horizontal gene transfer resulting origin of new ARB in environment.
From the records of the history of antibiotic use by the veterinary physicians of CCBDF, it was also known that they commonly used aminoglycosides antibiotics to treat cattle diseases (Table 2). It was interesting that wastewater samples collected from almost all of the locations of the sewerage lines of CCBDF showed the highest % of resistant bacteria to be against streptomycin, the most commonly used aminoglycosides at CCBDF. From the records, we also found that tuberculosis (TB) was the most common cattle disease at CCBDF and streptomycin was one of the most widely used aminoglycosides used for the treatment of TB as well as many other cattle diseases at CCBDF in the recovery shed. Therefore, it was expected that the bacterial isolates might show maximum resistance against streptomycin.
During bacteriological enumeration, resistant colonies were observed in all the plates containing waste-water samples. However, we did not find any such resistant colony in the plate for all the three aminoglycoside antibiotics in a water sample collected from the pond where no flow of cattle waste could reach. This finding strongly suggests that CCBDF wastes disposed to the sewerage lines contribute to the emergence of resistant bacteria in environment of the cattle farm.
Resistance to antibiotics, whether it arises by mutations or by conjugative transfer of R plasmids, is a serious problem for the treatment of cattle bacterial infections. Emergence of antibiotic resistance in microorganisms and its’ transfer horizontally and/or vertically limits the use of common antibiotics in the control of infectious diseases. Hence, great efforts are being made to understand the mechanisms involved in resistance and how to prevent its occurrence. The findings presented in this research work clearly demonstrate the positive correlation between waste-water and emergence of antibiotic resistance in environment. This result has notable biomedical importance to cattle as well public health. So far we know this is the first report in Bangladesh that demonstrates the possible role of cattle farm wastes in the pollution of environment with ARB and resistant gene pools. The scenario is plausible, but the risks cannot be estimated because of limited data availability. Hence, more extensive research is urgently needed to determine the environmental impacts of widely used antibiotics in medicine and agriculture on natural microbial communities.
REFERENCES
- Aarestrup, F.M. (1999) Association between the consumption of antimicrobial agents in animal husbandry and the occurrence of resistant bacteria among food animals. International Journal of Antimicrobial Agents, 12, 279-285. doi:10.1016/S0924-8579(99)90059-6
- Barbosa, T. and Levy, S.B. (2000b) The impact of antibiotic use on resistance development and persistence. Drug Resistance Updates, 3, 303-311. doi:10.1054/drup.2000.0167
- Bradford, P.A., Peterson, P.J., Fingerman, I.M. and White, D.G. (1999) Characterization of expanded cephalosporin resistance in E. coli isolates associated with bovine calf diarrhoeal disease. Journal of Antimicrobial Chemotherapy, 44, 607- 610. doi:10.1093/jac/44.5.607
- Davies, J.E., Roberts, M.C., Levy, S.B., Miller, G.H., O’Brien, T.F. and Tenover, T.C. (1999) Antimicrobial resistance: An ecological perspective. Report from colloquium held in San Juan, Puerto Rico. American Academy of Microbiology, Washington.
- Paterson, D.L. (2002) Serious infections caused by enteric gram-negative bacilli—Mechanisms of antibiotic resistance and implications for therapy of gram-negative sepsis in the transplanted patient. Seminars in Respiratory Infections, 17, 260-264. doi:10.1053/srin.2002.36446
- Rupp, M.E. and Fey, P.D. (2003) Extended spectrum ß-lactamase (ESBL)-producing Enterobacteriaceae: Considerations for diagnosis, prevention and drug treatment. Drugs, 63, 353-365. doi:10.2165/00003495-200363040-00002
- Hanzawa, Y., Oka, C., Ishiguru, N. and Sato, G. (1984) Antibiotic resistant coliforms in the waste of piggeries and dairy farms. Japanese Journal of Veterinary Science, 46, 363-372. doi:10.1292/jvms1939.46.363
- Owens, W.E., Nickerson, S.C., Boddie, R.L., Tomita, G.M. and Ray, C.H. (2001) Prevalence of mastitis in dairy heifers and effectiveness of antibiotic therapy. Journal of Dairy Science, 84, 814-817. doi:10.3168/jds.S0022-0302(01)74538-9
- Van Den Bogaard, A.E. and Stobberingh, E.E. (1999) Antibiotic usage in animals: Impact on bacterial resistance and public health. Drugs, 58, 589-607. doi:10.2165/00003495-199958040-00002
- Winokur, P.L., Voinstein, D.L., Hoffman, L.J. and Doern, G.V. (2001) Evidence for transfer of CMY-2 AmpC ß- lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrobial Agents and Chemotherapy, 45, 2716-2722. doi:10.1128/AAC.45.10.2716-2722.2001
- Ferber, D. (2002) Livestock feed ban preserves drugs’ power. Science, 295, 27-28. doi:10.1126/science.295.5552.27a
- CLSI (2009) Performance standards for antimicrobial susceptibility testing. CLSI (Clinical and Laboratory Standards Institute), Wayne.
- John, P.H. (2005) Laboratory exercises in microbiology. McGraw-Hill, New York.
NOTES
*Corresponding author.

