Crystal Structure Theory and Applications
Vol.2 No.4(2013), Article ID:41108,4 pages DOI:10.4236/csta.2013.24021
Purification, Crystallization and Preliminary X-Ray Diffraction Analysis of Exodeoxyribonuclease III from Crenarchaeon Sulfolobus tokodaii Strain 7
1Faculty of Pharmaceutical Sciences, Sojo University, Kumamoto, Japan
2School of Pharmacy, Showa University, Tokyo, Japan
3Faculty of Pharmacy, Iwaki Meisei University, Iwaki, Japan
Email: *miyamoto@ph.sojo-u.ac.jp
Copyright © 2013 Shuichi Miyamoto et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received October 13, 2013; revised November 12, 2013; accepted December 6, 2013
Keywords: Crenarchaeon; Crystallization; Exodeoxyribonuclease; Sulfolobus tokodaii; X-Ray Diffraction
ABSTRACT
Exodeoxyribonuclease III (EXOIII) acts as a 3’→5’ exonuclease and is homologous to purinic/apyrimidinic (AP) endonuclease (APE), which plays an important role in the base excision repair pathway. To structurally investigate the reaction and substrate recognition mechanisms of EXOIII, a crystallographic study of EXOIII from Sulfolobus tokodaii strain 7 was carried out. The purified enzyme was crystallized by using the hanging-drop vapor-diffusion method. The crystals belonged to space group C2, with unit-cell parameters a = 154.2, b = 47.7, c = 92.4 Å, β = 125.8˚ and diffracted to 1.5 Å resolution.
1. Introduction
A variety of mechanisms exist to repair damaged DNA and maintain a high degree of genomic stability within cells. Endonucleases specific for abasic or apurinic/ apyrimidinic (AP) sites are a major component of the cellular DNA repair machinery. AP sites are generated by spontaneous base loss [1], direct action of reactive oxygen species [2] and damage-specific DNA glycosylases as repair intermediates in the DNA base excision repair (BER) pathway [3,4]. It is estimated that as many as 10,000 purine bases alone are lost in a mammalian cell each day [5]. The generated AP sites are potentially mutagenic and cytotoxic, because of the inhibition of DNA replication and transcription [6,7]. AP sites are specifically corrected by the BER pathway: AP endonucleases (APE) first recognize the AP site and then cleave the phosphodiester backbone immediately 5’ of the AP site, leaving a free 3’-hydroxyl nucleotide end and a deoxyribose 5’-phospate as termini [8,9].
Exodeoxyribonuclease III (EXOIII; EC 3.1.11.2) is homologous to APE and acts as a 3’→5’ exonuclease. It is considered that the AP site undergoes a unique conformational change upon protein binding that permits complex formation and activation of attacking water, leading to incision, in the presence of Mg2+ [10,11]. However, the detailed reaction and substrate recognition mechanisms of AP endonucleases are not fully understood yet. Although EXOIII and APE belong to the same family, they have different substrate specificities. The three dimensional structures of EXOIII from Escherichia coli (E. coli) [12] and human APE [13] have been elucidated, but structures of these two enzymes from the same species have yet to be determined. To reveal the structural basis for the reaction and substrate recognition mechanisms of these enzymes from the same organism, we crystallized EXOIII and APE from the crenarchaeon, Sulfolobus tokodaii strain 7 [14]. Sulfolobus tokodaii is an aerobic thermoacidophilic crenarchaeon that optimally grows at 353 K and at low pH. Therefore, we also hope to obtain structural insights into the thermostability of these enzymes by comparing their structures with those of corresponding enzymes from other mesophilic organisms.
Here, we report the expression, purification, crystallization and preliminary X-ray diffraction analysis of EXOIII from Sulfolobus tokodaii strain 7, that is stoEXOIII. The enzyme consists of 241 amino acids (NCBI reference code NP_377894; UniProt ID F9VNP2), and has a molecular weight of 28.08 kDa and an isoelectric point of 8.72.
2. Materials and Methods
All chemicals were special grade reagents purchased from Wako Pure Chemical Industries, Japan unless stated otherwise.
2.1. Expression and Purification
The stoEXOIII protein was expressed in E. coli Rosettagami (DE3) using the pET-11a expression system (Merck KGaA, Germany). Since neither affinity tag nor extra residue was included, the sequence of the expressed protein was the same as that of the database entry. Bacterial clones were cultivated at 310 K in Luria-Bertani medium (Sigma-Aldrich, USA) with 100 μg∙ml−1 ampicillin and 30 μg∙ml−1 chloramphenicol. The cells were harvested by centrifugation at 8000 g, resuspended in 20 mM Tris-HCl pH 8.0, 50 mM NaCl and sonicated on ice. The mixture was heated at 343 K for 30 min and the soluble fraction was collected by centrifugation at 8000 g for 15 min. Ammonium sulfate was added to the supernatant to 50% saturation and the mixture was centrifuged at 10,000 g for 15 min. Ammonium sulfate was again added to the supernatant, to a final concentration of 60% saturation. After centrifugation at 10,000 g for 15 min, the pellet was dissolved in 50 mM sodium phosphate buffer (pH = 7.0) and dialyzed against the same buffer. The desalted mixture was loaded onto a SP-Sepharose column (1 ml; GE Healthcare Life Sciences) pre-equilibrated with 50 mM sodium phosphate buffer (pH = 7.0). The protein was eluted with a linear gradient of 0 - 1 M NaCl in the same buffer and protein purity was confirmed by SDS-PAGE. Simultaneous removal of NaCl and concentration of the protein was achieved by several rounds of diafiltration using an Amicon Ultra-10 Centrifugal Filter device (Millipore) with 20 mM sodium phosphate buffer (pH = 7.0) to a final protein concentration of 10 mg∙ml−1. The final yield of pure protein was around 0.9 mg per liter of culture.
2.2. Crystallization
Initial crystallization conditions were screened by the hanging-drop vapour-diffusion technique at 293 K using Index, Crystal Screen and Crystal Screen 2 crystal screening kits (Hampton Research, USA). The crystallization droplets were prepared on siliconized cover slides by mixing 1 μl protein solution with 1 μl reservoir solution; the drops were then equilibrated against 800 μl of reservoir solution. Several single crystals appeared within a few days in several solutions containing PEG 4000 - 8000 (1st grade, Wako, Japan) as precipitant. Crystallization conditions were further optimized manually by refinement of buffer choice and PEG concentration. The quality of the crystals were checked from diffraction images obtained using an R-AXIS IV++ X-ray diffractometer (Rigaku, Japan). Finally, the most suitable precipitant solution for crystallization was 15% (w/v) PEG 6000, 0.1 M Bis-Tris pH 5.5 or 0.1 M HEPES pH 7.0, and 0.1 M L(-)-proline. stoEXOIII protein crystals with approximate dimensions of 1.0 × 0.1 × 0.1 mm were obtained in about one week at 293 K (Figure 1).
2.3. Data Collection
A stoEXOIII crystal was picked from the crystallization droplet using a mounted nylon loop and placed directly into a cold nitrogen-gas stream at 100 K. X-ray diffraction data were collected with a CCD detector (Quantum 210, Area Detector Systems Co., USA) on beamline BLNW12 of the Photon Factory-Advanced Ring (PF-AR), Tsukuba, Japan, using a wavelength of 1.00 Å. The distance between the crystal and detector was 110 mm. A total of 360 images with 0.5 degrees oscillation were collected with an exposure time of 10 seconds.
3. Results and Discussion
Diffraction data were obtained in the resolution range 100 - 1.5 Å (Figure 2) and were then indexed and scaled using DENZO and SCALEPACK from the HKL-2000 program suite [15]. The crystals belong to the monoclinic space group C2, with unit-cell parameters a = 154.2, b = 47.7, c = 92.4 Å, β = 125.8˚. A total of 87532 independent reflections obtained with a multiplicity of 5.1 at 1.5 Å resolution reached 99.3% completeness with an Rmerge of 6.7%. Assuming that two monomers are present in the asymmetric unit, the Matthews coefficient was calculated to be 2.4 Å3∙Da−1 [16]. This corresponds to an estimated

Figure 1. Crystals of stoEXOIII grown in 15% (w/v) PEG 6000, 0.1 M HEPES pH 7.0 and 0.1 M L(-)-proline using the hanging-drop vapor-diffusion method at 293 K. The size of the main crystal is approximately 1.0 × 0.1 × 0.1 mm.
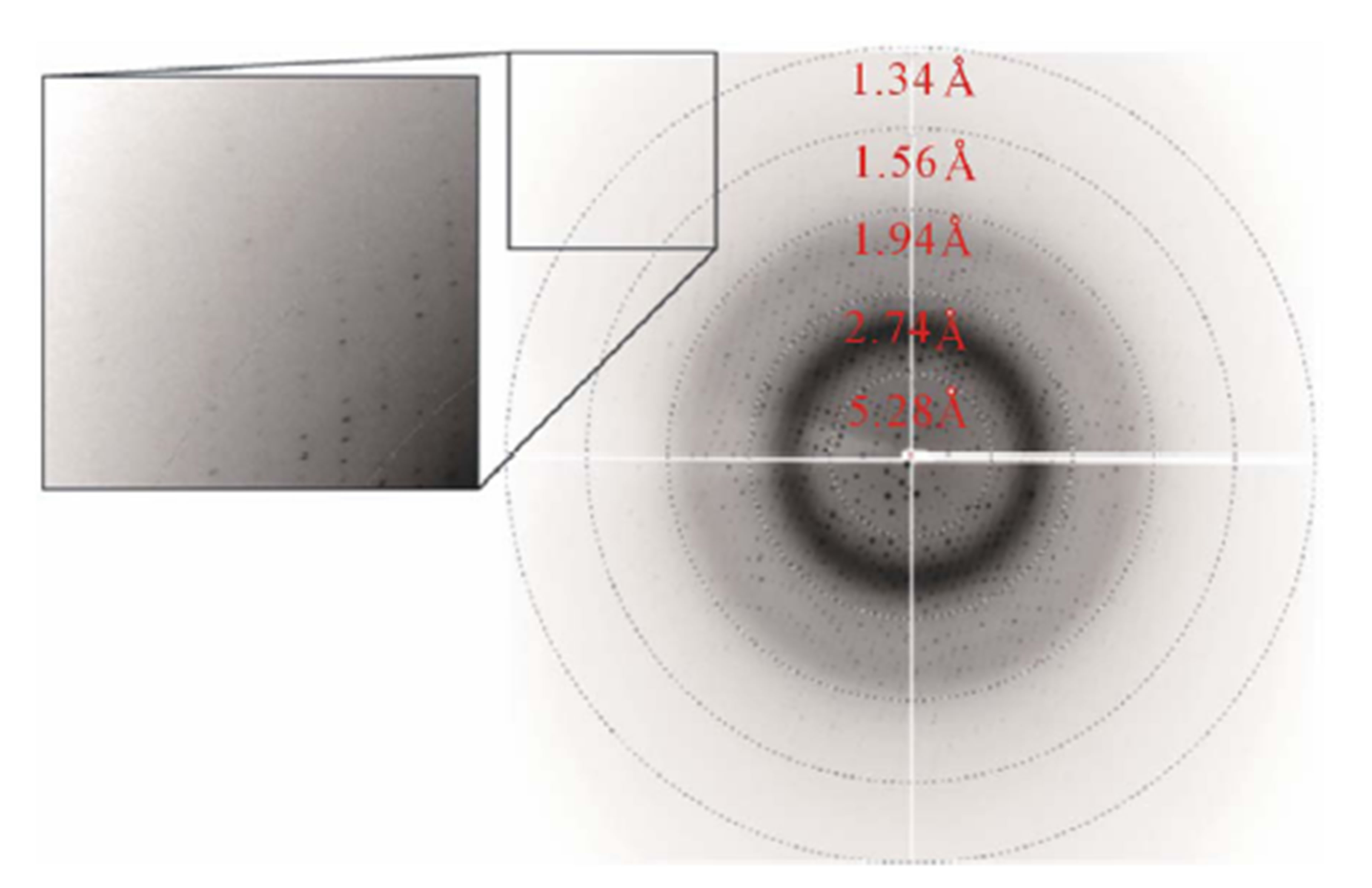
Figure 2. X-ray diffraction image. A corner section at the upper left is enlarged and shown in the inset.

Table 1. Data-collection statistics. Values in parentheses are for the highest resolution shell.
solvent content of 49%, which is in the range normally observed for protein crystals. The relevant data-collection statistics are summarized in Table 1. The crystal structure determination of stoEXOIII is under way by the molecular replacement method using the MOLREP program [17] in the CCP4 suite [18] at 1.5 Å resolution.
4. Conclusion
Exodeoxyribonuclease III from the crenarchaeon, Sulfolobus tokodaii strain 7, was purified and then crystallized by using the hanging-drop vapor-diffusion method. The crystals belonged to space group C2, with unit-cell parameters a = 154.2, b = 47.7, c = 92.4 Å, β = 125.8˚ and diffracted to 1.5 Å resolution.
5. Acknowledgements
We are grateful to the scientists and staff at the Photon Factory for use of the synchrotron. This work was partly supported by the National Project on Protein Structural and Functional Analysis by the Ministry of Education, Culture, Sports, Science and Technology of Japan (Protein 3000 project). We thank Professor Seiki Kuramitsu for organizing the research group in the program.
REFERENCES
- T. Lindahl, “Instability and Decay of the Primary Structure of DNA,” Nature, Vol. 362, No. 6422, 1993, pp. 709-715. http://dx.doi.org/10.1038/362709a0
- B. Demple and L. Harrison, “Repair of Oxidative Damage to DNA: Enzymology and Biology,” Annual Review of Biochemistry, 1994, Vol. 63, 1994, pp. 915-948.
- H. E. Krokan, R. Standal and G. Slupphang, “DNA Glycosylases in the Base Excision Repair of DNA,” Biochemical Journal, 1997, Vol. 325, pp. 1-16.
- J. Baute and A. Depicker, “Base Excision Repair and Its Role in Maintaining Genome Stability,” Critical Reviews in Biochemistry and Molecular Biology, Vol. 43, No. 4, 2008, pp. 239-276. http://dx.doi.org/10.1080/10409230802309905
- T. Lindahl and B. Nyberg, “Rate of Depurination of Native Deoxyribonucleic Acid,” Biochemistry, Vol. 11, No. 19, 1972, pp. 3610-3618. http://dx.doi.org/10.1021/bi00769a018
- L. A. Loeb and B. D. Preston, “Mutagenesis by Apurinic/ Apyrimidinic Sites,” Annual Review of Genetics, Vol. 20, 1986, pp. 201-230. http://dx.doi.org/10.1146/annurev.ge.20.120186.001221
- S. L. Yu, S. K. Lee, R. E. Johnson, L. Prakash and S. Prakash, “The Stalling of Transcription at Abasic Sites Is Highly Mutagenic,” Molecular and Cellular Biology, Vol. 23, No. 1, 2003, pp. 382-388. http://dx.doi.org/10.1128/MCB.23.1.382-388.2003
- P. W. Doetsch and R. P. Cunningham, “The Enzymology of Apurinic/Apyrimidinic Endonucleases,” Mutation Research, Vol. 236, No. 2-3, 1990, pp. 173-201. http://dx.doi.org/10.1016/0921-8777(90)90004-O
- G. Barzilay and I. D. Hickson, “Structure and Function of Apurinic/Apyrimidinic Endonucleases,” Bioessays, Vol. 17, No. 8, 1995, pp. 713-719. http://dx.doi.org/10.1002/bies.950170808
- J. P. Erzberger, D. Barsky, O. D. Schärer, M. E. Colvin and D. M. Wilson III, “Elements in Abasic Site Recognition by the Major Human and Escherichia coli Apurinic/ Apyrimidinic Endonucleases,” Nucleic Acids Research, Vol. 26, No. 11, 1998, pp. 2771-2778. http://dx.doi.org/10.1093/nar/26.11.2771
- D. Lu, J. Silhan, J. T. MacDonald, E. P. Carpenter, K. Jensen, C. M. Tang, G. S. Baldwin and P. S. Freemont, “Structural Basis for the Recognition and Cleavage of Abasic DNA in Neisseria meningitidis,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 109, No. 42, 2012, pp. 16852-16857. http://dx.doi.org/10.1073/pnas.1206563109
- C. D. Mol, C.-F. Kuo, M. M. Thayer, R. P. Cunningham and J. A. Tainer, “Structure and Function of the Multifunctional DNA-Repair Enzyme Exonuclease III,” Nature, Vol. 374, No. 6520, 1995, pp. 381-386. http://dx.doi.org/10.1038/374381a0
- M. A. Gorman, S. Morera, D. G. Rothwell, E. de La Fortelle, C. D. Mol, J. A. Tainer, I. D. Hickson and P. S. Freemont, “The Crystal Structure of the Human DNA Repair Endonuclease HAP1 Suggests the Recognition of Extra-Helical Deoxyribose at DNA Abasic Sites,” EMBO Journal, Vol. 16, No. 21, 1997, pp. 6548-6558. http://dx.doi.org/10.1093/emboj/16.21.6548
- Y. Kawarabayasi, Y. Hino, H. Horikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Kato, T. Yoshizawa, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, S. Masuda, M. Yanagii, M. Nishimura, A. Yamagishi, T. Oshima and H. Kikuchi, “Complete Genome Sequence of an Aerobic Thermoacidophilic Crenarchaeon, Sulfolobus tokodaii Strain 7,” DNA Research, Vol. 8, No. 4, 2001, pp. 123-140. http://dx.doi.org/10.1093/dnares/8.4.123
- Z. Otwinowski and W. Minor, “Processing of X-Ray Diffraction Data Collected in Oscillation Mode,” Methods in Enzymology, Vol. 276, 1997, pp. 307-326. http://dx.doi.org/10.1016/S0076-6879(97)76066-X
- B. W. Matthews, “Solvent Content of Protein Crystals,” Journal of Molecular Biology, Vol. 33, No. 2, 1968, pp. 491-497. http://dx.doi.org/10.1016/0022-2836(68)90205-2
- A. Vagin, and A. Teplyakov, “MOLREP: An Automated Program for Molecular Replacement,” Journal of Applied Crystallography, Vol. 30, Part 10, 1997, pp. 1022-1025. http://dx.doi.org/10.1107/S0021889897006766
- Collaborative Computational Project, Number 4, “The CCP4 Suite: Programs for Protein Crystallography,” Acta Crystallographica Section D, Vol. 50, Part 5, 1994, pp. 760-763. http://dx.doi.org/10.1107/S0907444994003112
NOTES
*Corresponding author.

